Gene
KWMTBOMO00164
Annotation
putative_RNA-directed_DNA_polymerase_from_transposon_BS-like_Protein_[Tribolium_castaneum]
Location in the cell
Mitochondrial Reliability : 1.745 PlasmaMembrane Reliability : 1.244
Sequence
CDS
ATGGAGACAGCGTATAAAAAATATTTGTTGAGTATAGAGAGAGACATCGGTGATAATCCAAAAATATTTTGGAAGTTTATTAAAGATAAGAGACAAAGTAGGCATAATTGTAATAAATATTATGATTACTTGGGAAAACCTGTGGAAGGACAATACGCGGCAGATGCATTTGCAACTTACTTCAGTTCAGTTTTCTTAGCCGATGAACCCAAATTAGATCCAACTGAGGCCCAGCATAGTGCAGCAATAACATCACAGTTTTGTTCCAGTGCTCGAGTAGCTTTATTGACTGTAGATAATTCCGACCTGCGCTGTGCTGTGAAGCGCATTAAGCCGTTCTCCGCATCAGGTCCAGATCGTATCCCTTCCTTTTTGGCTAAGGATTGTATAAGTGTGCTTAGTGTACCACTACTACATATTTATAATCTCTCACTCCAAATGTCCACATATCCATCTACGTGGAAGCACACTAGAGTAACTCCGGTCTTCAAGGGTGGTGACCGCACAGATGTGAAAAACTATCGACCCATCGCAGTGTTATCAGTATTTGCAAAGATATTTGAGTCCATTATTAATAAACATGTTAGTGCGCAGATCCGGAACCAATTGACTGACAGTCAACATGGTTTTCGAGCTGGTAGATCTACGGCAACTAACCATATTAATTTCGTCAACTATGTTGCTGCTGAAATGGATGCAGGGAAACAAGTCGACGCTGTATACTTAGACTTTGAAAAAGCCTTCGACCGTGTCGATAATGACGTACTGTTGATGAAATTTGCTTCATTCGGATTTACCCCAAAACTCCTCAAATTATTTTCTTCTTATCTCTCTGATAGGTCTCAGCATGTTGAGTTAGCGGGTTTCAAATCTAAAACTTATTTTACAAGATCAGGGGTCAGTCAGGGTAGTACGCTTGGTCCCACTCAGTTTTTAATCATGATAAATGATCTATCTTCTATTCTCAGTGAGAGGACCAAGTGTCTTATGTTTGCGGACGATCTTAAGCTGTACGTTGGGGTCGGGTGTGATGCTGAGCACCATGCTTTGCAACGCGACATTACTCGGGTTGCAGAATGGGGAGAGCGTAACCGCTTGCATTTTAATACTAATAAATGCAAGGTCGTTGCTTACAGCCGGTCACGGTCGCCCGTGTTGTTTCCATACCATTTATCTGGAGTCATTTTGAATCATGTGGCAGTTATACGTGACCTGGGAGTTATTTTCGATTCCAAGCTAACGTTTAAAGATCACATACAAAATATATGTAAAAGGGGTAGTGGTCTGCTCGGGTTCATAATTCGTCAGACGCACGACTTTGTAGGCCATCGTGTGATTGTTATGCTATACGATGCCTATTTGCGAAGTAGTCTTGAATATAACGCACTCGTGTGGGATCCTCGCGAAGCAGTTCAAAAGCTAATGCTGGAAAAACTACATAAAAAATTTTGCAGGTTCCTATTCAAGAAAATGTATGGATACTATCCGTTCTTGTATCCATCACTTTATGTGACTGGTATGGTCGGATTAGATACTCTTGAGCTCAGACGGAAGATGACTCTTATGGTGCATTATTATCAGCTATTGCATAACAAAATTGATAACTCCACTGCACTGGAATCAGTGTCGTTATATGTGCCTGATGGCTACATGCGCGGATGTATGTGTGGAGGGGTGCGTCGCCAGCGGCAGCTTCTTAGTACTATGCACGCGCGTACGCAGCACACCGCTAACTCCCCGACGCATCGTGCTGTCTCGTTACTCAATGGCTTGCTAGCCTATGCTCCGGAAATGGATATATTCCATGATGGTTTGCAGCGATTTATTGTTGTTGTTAGAAATTACTTAAGCTAA
Protein
METAYKKYLLSIERDIGDNPKIFWKFIKDKRQSRHNCNKYYDYLGKPVEGQYAADAFATYFSSVFLADEPKLDPTEAQHSAAITSQFCSSARVALLTVDNSDLRCAVKRIKPFSASGPDRIPSFLAKDCISVLSVPLLHIYNLSLQMSTYPSTWKHTRVTPVFKGGDRTDVKNYRPIAVLSVFAKIFESIINKHVSAQIRNQLTDSQHGFRAGRSTATNHINFVNYVAAEMDAGKQVDAVYLDFEKAFDRVDNDVLLMKFASFGFTPKLLKLFSSYLSDRSQHVELAGFKSKTYFTRSGVSQGSTLGPTQFLIMINDLSSILSERTKCLMFADDLKLYVGVGCDAEHHALQRDITRVAEWGERNRLHFNTNKCKVVAYSRSRSPVLFPYHLSGVILNHVAVIRDLGVIFDSKLTFKDHIQNICKRGSGLLGFIIRQTHDFVGHRVIVMLYDAYLRSSLEYNALVWDPREAVQKLMLEKLHKKFCRFLFKKMYGYYPFLYPSLYVTGMVGLDTLELRRKMTLMVHYYQLLHNKIDNSTALESVSLYVPDGYMRGCMCGGVRRQRQLLSTMHARTQHTANSPTHRAVSLLNGLLAYAPEMDIFHDGLQRFIVVVRNYLS
Summary
Uniprot
A0A2W1B2I7
A0A2W1BVH6
A0A2W1BAZ4
A0A2A4JV67
A0A1B6KA64
A0A1B6KI85
+ More
A0A1Y1MD08 D7GXK5 D7GXK8 D7GXK7 V5GTW8 A0A1Y1KXC2 D7GXK2 A0A1Y1KS26 A0A1Y1NC99 A0A1Y1JXM0 A0A0R3Q9H7 A0A3P7SV72 D7GXK9 A0A1Y1MYJ6 A0A1Y1K5G1 A0A147BQA9 A0A147BKN6 A0A147BMS6 A0A023EYX7 A0A147BKW8 J9LK97 A0A147BPQ9 A0A147BLW3 A0A139W9M8 A0A1Y1NIA6 A0A2S2P7I8 A0A147BM65 A0A0R3Q7A0 A0A2A4JB85 A0A2S2P1K1 A0A1W7R6M7 A0A1W7R6F5 A0A0P6JS06 A0A131XS42 A0A2J7PBJ9 A0A2J7Q1D2 A0A0P6IVX2 A0A2J7Q308 A0A0P6J4T9 A0A2S2NJ55 A0A131XS34 A0A1Y1MYU4 A0A224XJM9 A0A2S2P8J1 A0A2S2PKE0 A0A147BLG5 A0A131XUY4 J9JJD6 A0A0P6IZ62 A0A2J7PH98 Q17003 F5HRJ0 A0A2J7QB70 A0A2J7Q4R1 A0A2J7Q8V3 A0A2J7PTY4 A0A2H8TY56 A0A131XPZ6 U5ETF0 A0A1G4M228 A0A2J7PUN5 A0A2J7Q6G3 A0A147BJQ2 A0A3P7E401 A0A2M4CUU6 X1X2L3 A0A2J7QVC0 A0A2M4CUL1 A0A147BSB2 A0A2J7PW08 A0A131XV73 X1WVM0 A0A1W7R687 A0A0K8VXH1 A0A131XR06 A0A0A1X4N6 A0A2J7R0I0 A0A146KUF2 J9LUC1 X1WLK7 A0A1W7R665 A0A131XTD9 A0A2S2P7T9 X1WZG3 A0A146MAH9 J9L2B5 A0A2S2P3W2 J9KYJ0 A0A147BI04 A0A1Y1MYW8 V5H8T0
A0A1Y1MD08 D7GXK5 D7GXK8 D7GXK7 V5GTW8 A0A1Y1KXC2 D7GXK2 A0A1Y1KS26 A0A1Y1NC99 A0A1Y1JXM0 A0A0R3Q9H7 A0A3P7SV72 D7GXK9 A0A1Y1MYJ6 A0A1Y1K5G1 A0A147BQA9 A0A147BKN6 A0A147BMS6 A0A023EYX7 A0A147BKW8 J9LK97 A0A147BPQ9 A0A147BLW3 A0A139W9M8 A0A1Y1NIA6 A0A2S2P7I8 A0A147BM65 A0A0R3Q7A0 A0A2A4JB85 A0A2S2P1K1 A0A1W7R6M7 A0A1W7R6F5 A0A0P6JS06 A0A131XS42 A0A2J7PBJ9 A0A2J7Q1D2 A0A0P6IVX2 A0A2J7Q308 A0A0P6J4T9 A0A2S2NJ55 A0A131XS34 A0A1Y1MYU4 A0A224XJM9 A0A2S2P8J1 A0A2S2PKE0 A0A147BLG5 A0A131XUY4 J9JJD6 A0A0P6IZ62 A0A2J7PH98 Q17003 F5HRJ0 A0A2J7QB70 A0A2J7Q4R1 A0A2J7Q8V3 A0A2J7PTY4 A0A2H8TY56 A0A131XPZ6 U5ETF0 A0A1G4M228 A0A2J7PUN5 A0A2J7Q6G3 A0A147BJQ2 A0A3P7E401 A0A2M4CUU6 X1X2L3 A0A2J7QVC0 A0A2M4CUL1 A0A147BSB2 A0A2J7PW08 A0A131XV73 X1WVM0 A0A1W7R687 A0A0K8VXH1 A0A131XR06 A0A0A1X4N6 A0A2J7R0I0 A0A146KUF2 J9LUC1 X1WLK7 A0A1W7R665 A0A131XTD9 A0A2S2P7T9 X1WZG3 A0A146MAH9 J9L2B5 A0A2S2P3W2 J9KYJ0 A0A147BI04 A0A1Y1MYW8 V5H8T0
Pubmed
EMBL
KZ150413
PZC70972.1
KZ149924
PZC77634.1
KZ150173
PZC72589.1
+ More
NWSH01000614 PCG75292.1 GEBQ01031644 JAT08333.1 GEBQ01028810 JAT11167.1 GEZM01038166 JAV81746.1 GG694237 EFA13480.1 GG694238 EFA13486.1 EFA13487.1 GALX01004783 JAB63683.1 GEZM01073829 GEZM01073824 GEZM01073823 JAV64810.1 EFA13482.1 GEZM01075215 JAV64203.1 GEZM01007159 JAV95329.1 GEZM01097911 JAV54049.1 UZAG01001867 VDO12276.1 EFA13485.1 GEZM01017372 JAV90742.1 GEZM01096738 GEZM01096736 GEZM01096735 GEZM01096734 JAV54606.1 GEGO01002809 JAR92595.1 GEGO01004110 JAR91294.1 GEGO01003622 JAR91782.1 GBBI01004294 JAC14418.1 GEGO01004033 JAR91371.1 ABLF02022753 GEGO01002670 JAR92734.1 GEGO01003658 JAR91746.1 KQ972212 KYB24594.1 GEZM01001798 JAV97519.1 GGMR01012775 MBY25394.1 GEGO01003558 JAR91846.1 UZAG01001120 VDO10445.1 NWSH01002032 PCG69367.1 GGMR01010742 MBY23361.1 GEHC01000920 JAV46725.1 GEHC01000932 JAV46713.1 GDUN01000797 JAN95122.1 GEFM01006319 JAP69477.1 NEVH01027086 PNF13710.1 NEVH01019411 PNF22402.1 GDUN01000798 JAN95121.1 NEVH01019072 PNF22968.1 GDUN01000801 JAN95118.1 GGMR01004539 MBY17158.1 GEFM01006329 JAP69467.1 GEZM01017202 JAV90842.1 GFTR01008205 JAW08221.1 GGMR01013083 MBY25702.1 GGMR01016717 MBY29336.1 GEGO01003785 JAR91619.1 GEFM01006330 JAP69466.1 ABLF02028025 ABLF02028028 GDUN01000793 JAN95126.1 NEVH01025142 PNF15711.1 U03849 AAA53489.1 AB607192 BAK26811.1 NEVH01025296 NEVH01016302 NEVH01006564 PNF15439.1 PNF25825.1 PNF37995.1 NEVH01018377 PNF23575.1 NEVH01016945 PNF25017.1 NEVH01021219 PNF19791.1 GFXV01006926 MBW18731.1 GEFM01006322 JAP69474.1 GANO01004402 JAB55469.1 FLMD02000119 SCV65652.1 NEVH01021194 PNF20040.1 NEVH01017475 PNF24177.1 GEGO01004385 JAR91019.1 UYWW01005329 VDM14076.1 GGFL01004857 MBW69035.1 ABLF02022364 ABLF02041596 NEVH01010477 PNF32532.1 GGFL01004858 MBW69036.1 GEGO01001755 JAR93649.1 NEVH01020939 PNF20522.1 GEFM01006325 JAP69471.1 ABLF02006362 GEHC01000958 JAV46687.1 GDHF01008728 JAI43586.1 GEFM01006327 JAP69469.1 GBXI01008013 JAD06279.1 NEVH01008240 PNF34337.1 GDHC01020139 JAP98489.1 ABLF02014660 ABLF02019782 GEHC01000981 JAV46664.1 GEFM01006321 JAP69475.1 GGMR01012902 MBY25521.1 ABLF02041544 GDHC01002190 JAQ16439.1 ABLF02007491 ABLF02041729 GGMR01011552 MBY24171.1 ABLF02027115 ABLF02027120 ABLF02055498 GEGO01005479 JAR89925.1 GEZM01017203 JAV90839.1 GANP01010944 JAB73524.1
NWSH01000614 PCG75292.1 GEBQ01031644 JAT08333.1 GEBQ01028810 JAT11167.1 GEZM01038166 JAV81746.1 GG694237 EFA13480.1 GG694238 EFA13486.1 EFA13487.1 GALX01004783 JAB63683.1 GEZM01073829 GEZM01073824 GEZM01073823 JAV64810.1 EFA13482.1 GEZM01075215 JAV64203.1 GEZM01007159 JAV95329.1 GEZM01097911 JAV54049.1 UZAG01001867 VDO12276.1 EFA13485.1 GEZM01017372 JAV90742.1 GEZM01096738 GEZM01096736 GEZM01096735 GEZM01096734 JAV54606.1 GEGO01002809 JAR92595.1 GEGO01004110 JAR91294.1 GEGO01003622 JAR91782.1 GBBI01004294 JAC14418.1 GEGO01004033 JAR91371.1 ABLF02022753 GEGO01002670 JAR92734.1 GEGO01003658 JAR91746.1 KQ972212 KYB24594.1 GEZM01001798 JAV97519.1 GGMR01012775 MBY25394.1 GEGO01003558 JAR91846.1 UZAG01001120 VDO10445.1 NWSH01002032 PCG69367.1 GGMR01010742 MBY23361.1 GEHC01000920 JAV46725.1 GEHC01000932 JAV46713.1 GDUN01000797 JAN95122.1 GEFM01006319 JAP69477.1 NEVH01027086 PNF13710.1 NEVH01019411 PNF22402.1 GDUN01000798 JAN95121.1 NEVH01019072 PNF22968.1 GDUN01000801 JAN95118.1 GGMR01004539 MBY17158.1 GEFM01006329 JAP69467.1 GEZM01017202 JAV90842.1 GFTR01008205 JAW08221.1 GGMR01013083 MBY25702.1 GGMR01016717 MBY29336.1 GEGO01003785 JAR91619.1 GEFM01006330 JAP69466.1 ABLF02028025 ABLF02028028 GDUN01000793 JAN95126.1 NEVH01025142 PNF15711.1 U03849 AAA53489.1 AB607192 BAK26811.1 NEVH01025296 NEVH01016302 NEVH01006564 PNF15439.1 PNF25825.1 PNF37995.1 NEVH01018377 PNF23575.1 NEVH01016945 PNF25017.1 NEVH01021219 PNF19791.1 GFXV01006926 MBW18731.1 GEFM01006322 JAP69474.1 GANO01004402 JAB55469.1 FLMD02000119 SCV65652.1 NEVH01021194 PNF20040.1 NEVH01017475 PNF24177.1 GEGO01004385 JAR91019.1 UYWW01005329 VDM14076.1 GGFL01004857 MBW69035.1 ABLF02022364 ABLF02041596 NEVH01010477 PNF32532.1 GGFL01004858 MBW69036.1 GEGO01001755 JAR93649.1 NEVH01020939 PNF20522.1 GEFM01006325 JAP69471.1 ABLF02006362 GEHC01000958 JAV46687.1 GDHF01008728 JAI43586.1 GEFM01006327 JAP69469.1 GBXI01008013 JAD06279.1 NEVH01008240 PNF34337.1 GDHC01020139 JAP98489.1 ABLF02014660 ABLF02019782 GEHC01000981 JAV46664.1 GEFM01006321 JAP69475.1 GGMR01012902 MBY25521.1 ABLF02041544 GDHC01002190 JAQ16439.1 ABLF02007491 ABLF02041729 GGMR01011552 MBY24171.1 ABLF02027115 ABLF02027120 ABLF02055498 GEGO01005479 JAR89925.1 GEZM01017203 JAV90839.1 GANP01010944 JAB73524.1
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
A0A2W1B2I7
A0A2W1BVH6
A0A2W1BAZ4
A0A2A4JV67
A0A1B6KA64
A0A1B6KI85
+ More
A0A1Y1MD08 D7GXK5 D7GXK8 D7GXK7 V5GTW8 A0A1Y1KXC2 D7GXK2 A0A1Y1KS26 A0A1Y1NC99 A0A1Y1JXM0 A0A0R3Q9H7 A0A3P7SV72 D7GXK9 A0A1Y1MYJ6 A0A1Y1K5G1 A0A147BQA9 A0A147BKN6 A0A147BMS6 A0A023EYX7 A0A147BKW8 J9LK97 A0A147BPQ9 A0A147BLW3 A0A139W9M8 A0A1Y1NIA6 A0A2S2P7I8 A0A147BM65 A0A0R3Q7A0 A0A2A4JB85 A0A2S2P1K1 A0A1W7R6M7 A0A1W7R6F5 A0A0P6JS06 A0A131XS42 A0A2J7PBJ9 A0A2J7Q1D2 A0A0P6IVX2 A0A2J7Q308 A0A0P6J4T9 A0A2S2NJ55 A0A131XS34 A0A1Y1MYU4 A0A224XJM9 A0A2S2P8J1 A0A2S2PKE0 A0A147BLG5 A0A131XUY4 J9JJD6 A0A0P6IZ62 A0A2J7PH98 Q17003 F5HRJ0 A0A2J7QB70 A0A2J7Q4R1 A0A2J7Q8V3 A0A2J7PTY4 A0A2H8TY56 A0A131XPZ6 U5ETF0 A0A1G4M228 A0A2J7PUN5 A0A2J7Q6G3 A0A147BJQ2 A0A3P7E401 A0A2M4CUU6 X1X2L3 A0A2J7QVC0 A0A2M4CUL1 A0A147BSB2 A0A2J7PW08 A0A131XV73 X1WVM0 A0A1W7R687 A0A0K8VXH1 A0A131XR06 A0A0A1X4N6 A0A2J7R0I0 A0A146KUF2 J9LUC1 X1WLK7 A0A1W7R665 A0A131XTD9 A0A2S2P7T9 X1WZG3 A0A146MAH9 J9L2B5 A0A2S2P3W2 J9KYJ0 A0A147BI04 A0A1Y1MYW8 V5H8T0
A0A1Y1MD08 D7GXK5 D7GXK8 D7GXK7 V5GTW8 A0A1Y1KXC2 D7GXK2 A0A1Y1KS26 A0A1Y1NC99 A0A1Y1JXM0 A0A0R3Q9H7 A0A3P7SV72 D7GXK9 A0A1Y1MYJ6 A0A1Y1K5G1 A0A147BQA9 A0A147BKN6 A0A147BMS6 A0A023EYX7 A0A147BKW8 J9LK97 A0A147BPQ9 A0A147BLW3 A0A139W9M8 A0A1Y1NIA6 A0A2S2P7I8 A0A147BM65 A0A0R3Q7A0 A0A2A4JB85 A0A2S2P1K1 A0A1W7R6M7 A0A1W7R6F5 A0A0P6JS06 A0A131XS42 A0A2J7PBJ9 A0A2J7Q1D2 A0A0P6IVX2 A0A2J7Q308 A0A0P6J4T9 A0A2S2NJ55 A0A131XS34 A0A1Y1MYU4 A0A224XJM9 A0A2S2P8J1 A0A2S2PKE0 A0A147BLG5 A0A131XUY4 J9JJD6 A0A0P6IZ62 A0A2J7PH98 Q17003 F5HRJ0 A0A2J7QB70 A0A2J7Q4R1 A0A2J7Q8V3 A0A2J7PTY4 A0A2H8TY56 A0A131XPZ6 U5ETF0 A0A1G4M228 A0A2J7PUN5 A0A2J7Q6G3 A0A147BJQ2 A0A3P7E401 A0A2M4CUU6 X1X2L3 A0A2J7QVC0 A0A2M4CUL1 A0A147BSB2 A0A2J7PW08 A0A131XV73 X1WVM0 A0A1W7R687 A0A0K8VXH1 A0A131XR06 A0A0A1X4N6 A0A2J7R0I0 A0A146KUF2 J9LUC1 X1WLK7 A0A1W7R665 A0A131XTD9 A0A2S2P7T9 X1WZG3 A0A146MAH9 J9L2B5 A0A2S2P3W2 J9KYJ0 A0A147BI04 A0A1Y1MYW8 V5H8T0
PDB
6AR3
E-value=2.16507e-08,
Score=142
Ontologies
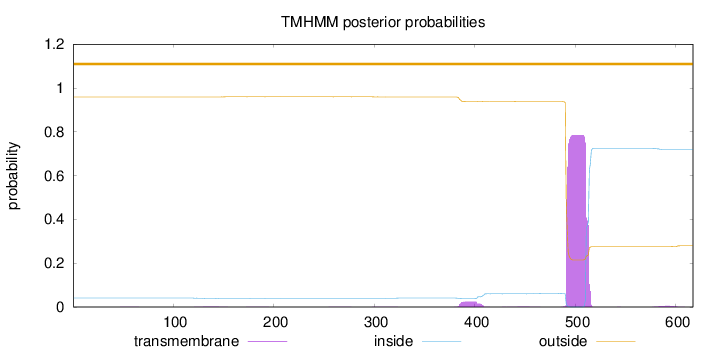
Topology
Length:
617
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
17.25357
Exp number, first 60 AAs:
0.00248
Total prob of N-in:
0.04072
outside
1 - 617
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
-0.720135
CLR
0
CSRT
0
Interpretation
Uncertain
