Gene
KWMTBOMO00156
Pre Gene Modal
BGIBMGA001996
Annotation
PREDICTED:_nervous_wreck_isoform_X1_[Bombyx_mori]
Full name
Protein nervous wreck
Location in the cell
Nuclear Reliability : 2.301
Sequence
CDS
ATGTGGAATGTGTGGCGCACGGTGCTGGAAGAGAACGAGAAGCTAGCACGCGCTCGTCTCGCAGCCGTCGAAGTGTTCCAACAGCACATCACCGATGAAGCGAAGCTGCTCCGACACCAGAAGCTCAGCTCTGCTAGGAAGTTTACGGAGGGCCTTGCTCAAGCCCACAAGGAGCTGCAGATGACTGTAGGTGAAGTTGACAAGACCAAGAAGCTGTACTTCGACGAGGAGCATACTGCTCATGACGTGCGCGACAAAGCTAAAGACATCGAGGAGAATAAACCGAGTAGCTCCCGGGGATCACCGAAACATCAACCGCCAGAAAGGAATGAACTAGTTGTAGATAAATATTGGTTGAAGAAGAAAAAAGGATCGTTCTTCCAATCGATTACTTCGCTGCAGAAGAACAGCGCCAAGGTTTCGTCCCGTCGTGATCAGCTGGAGGAGAAATCCACTGGCGCCAGGAACGATTATATTCTAAGCATTGCAGCTGCTAATGCTCACCAGAATCGCTACTTCCTAGTGGATCTTCAGACTTGCATGCAAAACATGGAGTCCTCGGTGTACGAGAAGGTTTCTGAGTACCTTACGCTTATGGGACGCACGGAACTCTTGACTTGCTCTGCCACCCAGCACTCATTTGGGAAGATCAGGGATCAAGCGCAGCAGCTGACCAGGGAATATAACTTGCAATGCTTGTATTTATATTATCCAGTGCTGAAGCAACATATCCAGTACGAGTTCGAGCCTTGCGACAATGATCCGATTGACACTATCACCATGGAGCACGAATCCGTGGCTGTCACTTTGGCTCAAGAGGCTCGCCGCTGGGCCACCCGCGTAGTTCGCGAGGCATCCCTCGTCCGGGATGCGAATCGCAAAATCAGTTCTTATCAAGCTATGCGGGAGTCTGGACATAAGGTGGACCCGAACGAACCCAACGGACCTGAGTTGGAAGTAAAGATGGATGAATTGCGTGCCACAATCCGTCGCTCTGAAATATCTAAGGCCAAGTACGAAGCTCGACTGGAATGCTTAAGAGTCGGAGGTGCTCCTGTAGATGATTGGCTAAAGGAAATCGATCTGCTGGCTGTCCAGGAGACACTGCCACGCAGCAGCAGCCTGCTAAGTGTTAGAACTGACGCATCCGGAGCCGCCGACCAGCCGAGCTCGGACTCTTTCTATGATAGCGACAACACTGAGGGTGAAGCAGCTGCTTCGGTGGCCGGCACCAGTGGCTCTGGTGGCCACCAACGTACCACTTCGGGCTCTCAGCATGAGGACGATCATGAAGATGAGGTTGACGCTGCCCTTGAGATTGAACGCCAGCGAATCGAGCAACTTGCGGGAGGATGGGATGATCCCACTCAAGTGAACTGGGGTGAATCCGACCCAGAGCCCTCTACTTCAGAGCCTATAGAACCACCAGCACCACCGCTTTACAAGTGTACTGCTCTTTACTCTTACACCGCTCAGAATCCGGATGAGTTGTCTATCATTGAGAACGAACAGCTTGAAGTGGTTGGAGAAGGCGATGGTGACGGTTGGCTGAAAGCTCGGAACTATCGCGGAGAAGAAGGCTACGTGCCACACAACTATCTCGATGTTGACCATGATCAGGCATCTAGCGCTCCAGGTTTGGTAAGCCAAATATCTTTCTCTTCTGTGGATTACACTGTGGAGGGTGAAGACGCTGACGTTGTCCAGTCACCGGATCAGATCTCTGTGATTTCGGCCCCCGTCGGCAAGCCTGATGAACCGACTAAAGCCGCAGAAGCAGCTCAGGGCGAGGCCGCTGCTGTAGCCCAGGGGCCACCAAAAGTCGACCTACCGACTCTCGGTTACTGCTTTGCTCTTTACGATTACGAAGCTGAGGCGTGCGATGAGCTCAACCTTGAAGAGGGACAGATTATTCGCGTTGTGTCCCGTAATGCTCATGATGTGGATGACGGTTGGTGGCGCGGAGAGACAAATGGCATTGTGGGAAATTTTCCGTCTCTCATTGTCGAGGAATGTGACGAGAATGGAGAACCACTGAGTGTCGTTGAAGAGGACTGGACCCCATCTGGATGCGCTCCTCCGGTCTTCGCTTCACCACCGACCTCACCACCAGGTGCTTCAGATGAAGCTGCGGTAAACGAGAGCAGCGCGGCCCCTCCTCTTGAACCACCACCGCCGCCGCCAGCTGATCTCGGTGACTCGATGGACAGCCAGCCCGACTTTAGCTTTAACCTAGAGTTGACCAGGAACCAGCAGGAGCAATACGGCATGCAGTTCTCGGAGCCCGCCGGCCCACCCGTGCCGCCCCCAGTCACTATTGTTGTTGATGAGGTTGCCAGCGATAATTTCGATGATGAGGACGAAGAAGAGATGCCGCAGCCGATGCCGGTGGTAAAGCCTGTGCCGCCGCCAGTTGAGCTGCCAAGTGCTGGCGAGTGCGGGCTGGGGGTCGCGCAGATTGTGATCACAGCGGCCACCCCGATGGTCGAGGAGCCGGAACTACCGTTCCCGCCCGCAGAGCCGCAGCCGCAGCCGCAACCGCAGATCGACGACGACGACGAGGAGTCGTCGCTGTCCGAGCAAACCGCGGTGTGCGTGCGTCCGCAGCCGACCTCTAGCTCCACAGGCTCGGAGGGCGATTCAACAGGCCCGCACTCTCCACCCGCTGCCCCACGAGCCGGTCGCGCCTCCATCCCCGATGAACTAGAGCCCGCCCAGCTGGCGAGACTCACCGACCTCAAGGAGTCCAACGCCTAA
Protein
MWNVWRTVLEENEKLARARLAAVEVFQQHITDEAKLLRHQKLSSARKFTEGLAQAHKELQMTVGEVDKTKKLYFDEEHTAHDVRDKAKDIEENKPSSSRGSPKHQPPERNELVVDKYWLKKKKGSFFQSITSLQKNSAKVSSRRDQLEEKSTGARNDYILSIAAANAHQNRYFLVDLQTCMQNMESSVYEKVSEYLTLMGRTELLTCSATQHSFGKIRDQAQQLTREYNLQCLYLYYPVLKQHIQYEFEPCDNDPIDTITMEHESVAVTLAQEARRWATRVVREASLVRDANRKISSYQAMRESGHKVDPNEPNGPELEVKMDELRATIRRSEISKAKYEARLECLRVGGAPVDDWLKEIDLLAVQETLPRSSSLLSVRTDASGAADQPSSDSFYDSDNTEGEAAASVAGTSGSGGHQRTTSGSQHEDDHEDEVDAALEIERQRIEQLAGGWDDPTQVNWGESDPEPSTSEPIEPPAPPLYKCTALYSYTAQNPDELSIIENEQLEVVGEGDGDGWLKARNYRGEEGYVPHNYLDVDHDQASSAPGLVSQISFSSVDYTVEGEDADVVQSPDQISVISAPVGKPDEPTKAAEAAQGEAAAVAQGPPKVDLPTLGYCFALYDYEAEACDELNLEEGQIIRVVSRNAHDVDDGWWRGETNGIVGNFPSLIVEECDENGEPLSVVEEDWTPSGCAPPVFASPPTSPPGASDEAAVNESSAAPPLEPPPPPPADLGDSMDSQPDFSFNLELTRNQQEQYGMQFSEPAGPPVPPPVTIVVDEVASDNFDDEDEEEMPQPMPVVKPVPPPVELPSAGECGLGVAQIVITAATPMVEEPELPFPPAEPQPQPQPQIDDDDEESSLSEQTAVCVRPQPTSSSTGSEGDSTGPHSPPAAPRAGRASIPDELEPAQLARLTDLKESNA
Summary
Description
Adapter protein that provides a link between vesicular membrane traffic and the actin assembly machinery. Acts together with Cdc42 to stimulate actin nucleation mediated by WASp and the ARP2/3 complex (PubMed:18701694, PubMed:27601635). Binds to membranes enriched in phosphatidylinositol 4,5-bisphosphate and causes local membrane deformation (PubMed:23761074, PubMed:26686642). Required for normal structure and function of synapses at the neuromuscular junction (PubMed:18701694, PubMed:14980202, PubMed:18498733, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Plays a role in synaptic vesicle trafficking (PubMed:29568072). Required for the release of a normal number of synaptic vesicles per action potential (PubMed:26567222).
Subunit
Homodimer (Probable) (PubMed:27601635). Interacts (via SH3 domain 1) with WASp (PubMed:14980202, PubMed:18701694, PubMed:27601635). Interacts (via SH3 domain 1) with shi/dynamin (PubMed:18701694, PubMed:18498733). Interacts (via SH3 domain 2) with Dap160 (PubMed:18701694, PubMed:18498733, PubMed:26686642). Interacts (via F-BAR domain) with SH3PX1 (PubMed:26567222). Interacts (via SH3 domain 2) with Snx16 (PubMed:21464232). Identified in a complex with Syn and Syt1 (PubMed:29568072).
Keywords
Alternative splicing
Cell junction
Cell membrane
Cell projection
Complete proteome
Cytoplasmic vesicle
Endosome
Lipid-binding
Membrane
Reference proteome
Repeat
SH3 domain
Synapse
Feature
chain Protein nervous wreck
splice variant In isoform 3, isoform 4 and isoform 7.
splice variant In isoform 3, isoform 4 and isoform 7.
Uniprot
F2Z9J3
F2Z9J4
F2Z9J5
H9IXL3
A0A194Q4X9
A0A194QUY6
+ More
A0A212FHJ0 A0A2H1WKM9 A0A2A4JQQ7 A0A182M6T9 A0A182GKU6 A0A182WI63 A0A182R9B1 A0A084WN04 A0A182QAQ5 A0A182N7I7 A0A182PEX6 A0A1S4F8X0 D7EJ83 A0A182V5H8 A0A182YJI0 A0A182JYR8 A0A182UDC0 A0A182X244 A0A182FFG9 A0A182T341 A0A182I6A5 Q0IEP6 A0A182J2H7 W5JVR5 A0A182L436 N6UJW0 A0A1W4X4Q9 A0A1W4XEA4 A0A1W4XFH8 A0A1W4XF75 A0A1B0D741 B0WLT2 A0A0C9RVR1 U4TPJ7 W8B4G9 W8BEK3 W8B4G6 A0A034VUZ4 A0A034VXJ3 A0A2A3EFW9 A0A0M3QW65 A0A1B0G7J7 A0A1B0C2P2 A0A034VVX8 A0A034VWM8 V9IEE5 A0A0Q9XAM2 A0A0Q9XMJ2 A0A0Q9XM53 B4KWM5 A0A088ATW5 A0A1I8PXU3 A0A0L0CNP5 A0A0Q9WUM9 A0A1A9ZC18 A0A0Q9WHZ9 A0A1A9XQZ4 A0A1A9UP20 A0A0P8ZWB8 A0A0Q9WSV5 A0A0P8XW46 A0A0P8XW74 A0A0Q9WHH4 A0A0Q9WSZ6 A0A0M9A852 A0A0P8YIX5 A0A1L8DME3 A0A0P9AMA5 A0A0P8YC42 A0A0A1X6B3 A0A1Y1M0Z7 A0A1I8PXW8 A0A0Q9WUJ2 A0A1Y1M549 B3M8W8 X2JAU8-5 A0A026W4P8 B4LHG5 A0A0L7QSK4 A0A0Q5UHK9 A0A154PA73 X2JAU8-2 X2JAU8-6 A0A0Q5U3S5 E0VZW6 A0A0R1E0A2 A0A0Q5UHN1 A0A0R1DUU2 A0A0Q5U2S9 B4QMQ6 A0A0R1E0C3 B4HKL4 X2JAU8 B4N6Z5
A0A212FHJ0 A0A2H1WKM9 A0A2A4JQQ7 A0A182M6T9 A0A182GKU6 A0A182WI63 A0A182R9B1 A0A084WN04 A0A182QAQ5 A0A182N7I7 A0A182PEX6 A0A1S4F8X0 D7EJ83 A0A182V5H8 A0A182YJI0 A0A182JYR8 A0A182UDC0 A0A182X244 A0A182FFG9 A0A182T341 A0A182I6A5 Q0IEP6 A0A182J2H7 W5JVR5 A0A182L436 N6UJW0 A0A1W4X4Q9 A0A1W4XEA4 A0A1W4XFH8 A0A1W4XF75 A0A1B0D741 B0WLT2 A0A0C9RVR1 U4TPJ7 W8B4G9 W8BEK3 W8B4G6 A0A034VUZ4 A0A034VXJ3 A0A2A3EFW9 A0A0M3QW65 A0A1B0G7J7 A0A1B0C2P2 A0A034VVX8 A0A034VWM8 V9IEE5 A0A0Q9XAM2 A0A0Q9XMJ2 A0A0Q9XM53 B4KWM5 A0A088ATW5 A0A1I8PXU3 A0A0L0CNP5 A0A0Q9WUM9 A0A1A9ZC18 A0A0Q9WHZ9 A0A1A9XQZ4 A0A1A9UP20 A0A0P8ZWB8 A0A0Q9WSV5 A0A0P8XW46 A0A0P8XW74 A0A0Q9WHH4 A0A0Q9WSZ6 A0A0M9A852 A0A0P8YIX5 A0A1L8DME3 A0A0P9AMA5 A0A0P8YC42 A0A0A1X6B3 A0A1Y1M0Z7 A0A1I8PXW8 A0A0Q9WUJ2 A0A1Y1M549 B3M8W8 X2JAU8-5 A0A026W4P8 B4LHG5 A0A0L7QSK4 A0A0Q5UHK9 A0A154PA73 X2JAU8-2 X2JAU8-6 A0A0Q5U3S5 E0VZW6 A0A0R1E0A2 A0A0Q5UHN1 A0A0R1DUU2 A0A0Q5U2S9 B4QMQ6 A0A0R1E0C3 B4HKL4 X2JAU8 B4N6Z5
Pubmed
19121390
26354079
22118469
26483478
24438588
17510324
+ More
18362917 19820115 25244985 20920257 23761445 20966253 23537049 24495485 25348373 17994087 18057021 26108605 25830018 28004739 10731132 12537572 14980202 18701694 18498733 21464232 23761074 26686642 26567222 27601635 29568072 24508170 30249741 20566863 17550304
18362917 19820115 25244985 20920257 23761445 20966253 23537049 24495485 25348373 17994087 18057021 26108605 25830018 28004739 10731132 12537572 14980202 18701694 18498733 21464232 23761074 26686642 26567222 27601635 29568072 24508170 30249741 20566863 17550304
EMBL
AB600238
BAK22648.1
AB600239
BAK22649.1
AB600240
BAK22650.1
+ More
BABH01016014 BABH01016015 BABH01016016 KQ459463 KPJ00593.1 KQ461108 KPJ09129.1 AGBW02008503 OWR53202.1 ODYU01009248 SOQ53526.1 NWSH01000870 PCG73783.1 AXCM01004836 JXUM01014113 JXUM01014114 JXUM01014115 KQ560394 KXJ82642.1 ATLV01024543 KE525352 KFB51598.1 AXCN02000573 AXCN02000574 DS497698 EFA12613.1 APCN01003602 CH477558 CH477317 EAT39001.1 EAT43741.1 ADMH02000276 ETN67155.1 APGK01021192 KB740293 ENN80951.1 AJVK01012324 DS231990 EDS30660.1 GBYB01013050 JAG82817.1 KB630005 KB630385 ERL83334.1 ERL83576.1 GAMC01018369 JAB88186.1 GAMC01018371 JAB88184.1 GAMC01018374 JAB88181.1 GAKP01012718 JAC46234.1 GAKP01012719 JAC46233.1 KZ288255 PBC30683.1 CP012525 ALC43609.1 CCAG010022578 JXJN01024608 JXJN01024609 GAKP01012720 JAC46232.1 GAKP01012717 JAC46235.1 JR041711 AEY59498.1 CH933809 KRG05574.1 KRG05575.1 KRG05569.1 EDW17472.2 KRG05570.1 KRG05572.1 KRG05573.1 KRG05576.1 JRES01000128 KNC33940.1 CH940647 KRF83877.1 KRF83873.1 KRF83874.1 CH902618 KPU78938.1 KRF83876.1 KPU78934.1 KPU78941.1 KRF83875.1 KRF83870.1 KQ435724 KOX78426.1 KPU78935.1 KPU78936.1 KPU78942.1 GFDF01006456 JAV07628.1 KPU78933.1 KPU78939.1 GBXI01007468 JAD06824.1 GEZM01042991 JAV79409.1 KRF83872.1 GEZM01042992 JAV79405.1 EDV41119.2 AE014296 AHN58019.1 KK107419 QOIP01000003 EZA51055.1 RLU24467.1 EDW68495.2 KQ414758 KOC61529.1 CH954178 KQS43370.1 KQ434846 KZC08284.1 KQS43369.1 AAZO01006553 AAZO01006554 DS235854 EEB18922.1 CM000159 KRK00922.1 KQS43364.1 KQS43366.1 KRK00920.1 KRK00923.1 KQS43368.1 CM000363 EDX09810.1 KRK00916.1 CH480815 EDW40817.1 CH964168 EDW80134.2
BABH01016014 BABH01016015 BABH01016016 KQ459463 KPJ00593.1 KQ461108 KPJ09129.1 AGBW02008503 OWR53202.1 ODYU01009248 SOQ53526.1 NWSH01000870 PCG73783.1 AXCM01004836 JXUM01014113 JXUM01014114 JXUM01014115 KQ560394 KXJ82642.1 ATLV01024543 KE525352 KFB51598.1 AXCN02000573 AXCN02000574 DS497698 EFA12613.1 APCN01003602 CH477558 CH477317 EAT39001.1 EAT43741.1 ADMH02000276 ETN67155.1 APGK01021192 KB740293 ENN80951.1 AJVK01012324 DS231990 EDS30660.1 GBYB01013050 JAG82817.1 KB630005 KB630385 ERL83334.1 ERL83576.1 GAMC01018369 JAB88186.1 GAMC01018371 JAB88184.1 GAMC01018374 JAB88181.1 GAKP01012718 JAC46234.1 GAKP01012719 JAC46233.1 KZ288255 PBC30683.1 CP012525 ALC43609.1 CCAG010022578 JXJN01024608 JXJN01024609 GAKP01012720 JAC46232.1 GAKP01012717 JAC46235.1 JR041711 AEY59498.1 CH933809 KRG05574.1 KRG05575.1 KRG05569.1 EDW17472.2 KRG05570.1 KRG05572.1 KRG05573.1 KRG05576.1 JRES01000128 KNC33940.1 CH940647 KRF83877.1 KRF83873.1 KRF83874.1 CH902618 KPU78938.1 KRF83876.1 KPU78934.1 KPU78941.1 KRF83875.1 KRF83870.1 KQ435724 KOX78426.1 KPU78935.1 KPU78936.1 KPU78942.1 GFDF01006456 JAV07628.1 KPU78933.1 KPU78939.1 GBXI01007468 JAD06824.1 GEZM01042991 JAV79409.1 KRF83872.1 GEZM01042992 JAV79405.1 EDV41119.2 AE014296 AHN58019.1 KK107419 QOIP01000003 EZA51055.1 RLU24467.1 EDW68495.2 KQ414758 KOC61529.1 CH954178 KQS43370.1 KQ434846 KZC08284.1 KQS43369.1 AAZO01006553 AAZO01006554 DS235854 EEB18922.1 CM000159 KRK00922.1 KQS43364.1 KQS43366.1 KRK00920.1 KRK00923.1 KQS43368.1 CM000363 EDX09810.1 KRK00916.1 CH480815 EDW40817.1 CH964168 EDW80134.2
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000075883
+ More
UP000069940 UP000249989 UP000075920 UP000075900 UP000030765 UP000075886 UP000075884 UP000075885 UP000007266 UP000075903 UP000076408 UP000075881 UP000075902 UP000076407 UP000069272 UP000075901 UP000075840 UP000008820 UP000075880 UP000000673 UP000075882 UP000019118 UP000192223 UP000092462 UP000002320 UP000030742 UP000242457 UP000092553 UP000092444 UP000092460 UP000009192 UP000005203 UP000095300 UP000037069 UP000008792 UP000092445 UP000092443 UP000078200 UP000007801 UP000053105 UP000000803 UP000053097 UP000279307 UP000053825 UP000008711 UP000076502 UP000009046 UP000002282 UP000000304 UP000001292 UP000007798
UP000069940 UP000249989 UP000075920 UP000075900 UP000030765 UP000075886 UP000075884 UP000075885 UP000007266 UP000075903 UP000076408 UP000075881 UP000075902 UP000076407 UP000069272 UP000075901 UP000075840 UP000008820 UP000075880 UP000000673 UP000075882 UP000019118 UP000192223 UP000092462 UP000002320 UP000030742 UP000242457 UP000092553 UP000092444 UP000092460 UP000009192 UP000005203 UP000095300 UP000037069 UP000008792 UP000092445 UP000092443 UP000078200 UP000007801 UP000053105 UP000000803 UP000053097 UP000279307 UP000053825 UP000008711 UP000076502 UP000009046 UP000002282 UP000000304 UP000001292 UP000007798
Interpro
IPR035460
FCHSD_SH3_1
+ More
IPR036028 SH3-like_dom_sf
IPR001452 SH3_domain
IPR031160 F_BAR
IPR001060 FCH_dom
IPR027267 AH/BAR_dom_sf
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR011011 Znf_FYVE_PHD
IPR034732 EPHD
IPR019786 Zinc_finger_PHD-type_CS
IPR019542 Enhancer_polycomb-like_N
IPR036028 SH3-like_dom_sf
IPR001452 SH3_domain
IPR031160 F_BAR
IPR001060 FCH_dom
IPR027267 AH/BAR_dom_sf
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR011011 Znf_FYVE_PHD
IPR034732 EPHD
IPR019786 Zinc_finger_PHD-type_CS
IPR019542 Enhancer_polycomb-like_N
Gene 3D
ProteinModelPortal
F2Z9J3
F2Z9J4
F2Z9J5
H9IXL3
A0A194Q4X9
A0A194QUY6
+ More
A0A212FHJ0 A0A2H1WKM9 A0A2A4JQQ7 A0A182M6T9 A0A182GKU6 A0A182WI63 A0A182R9B1 A0A084WN04 A0A182QAQ5 A0A182N7I7 A0A182PEX6 A0A1S4F8X0 D7EJ83 A0A182V5H8 A0A182YJI0 A0A182JYR8 A0A182UDC0 A0A182X244 A0A182FFG9 A0A182T341 A0A182I6A5 Q0IEP6 A0A182J2H7 W5JVR5 A0A182L436 N6UJW0 A0A1W4X4Q9 A0A1W4XEA4 A0A1W4XFH8 A0A1W4XF75 A0A1B0D741 B0WLT2 A0A0C9RVR1 U4TPJ7 W8B4G9 W8BEK3 W8B4G6 A0A034VUZ4 A0A034VXJ3 A0A2A3EFW9 A0A0M3QW65 A0A1B0G7J7 A0A1B0C2P2 A0A034VVX8 A0A034VWM8 V9IEE5 A0A0Q9XAM2 A0A0Q9XMJ2 A0A0Q9XM53 B4KWM5 A0A088ATW5 A0A1I8PXU3 A0A0L0CNP5 A0A0Q9WUM9 A0A1A9ZC18 A0A0Q9WHZ9 A0A1A9XQZ4 A0A1A9UP20 A0A0P8ZWB8 A0A0Q9WSV5 A0A0P8XW46 A0A0P8XW74 A0A0Q9WHH4 A0A0Q9WSZ6 A0A0M9A852 A0A0P8YIX5 A0A1L8DME3 A0A0P9AMA5 A0A0P8YC42 A0A0A1X6B3 A0A1Y1M0Z7 A0A1I8PXW8 A0A0Q9WUJ2 A0A1Y1M549 B3M8W8 X2JAU8-5 A0A026W4P8 B4LHG5 A0A0L7QSK4 A0A0Q5UHK9 A0A154PA73 X2JAU8-2 X2JAU8-6 A0A0Q5U3S5 E0VZW6 A0A0R1E0A2 A0A0Q5UHN1 A0A0R1DUU2 A0A0Q5U2S9 B4QMQ6 A0A0R1E0C3 B4HKL4 X2JAU8 B4N6Z5
A0A212FHJ0 A0A2H1WKM9 A0A2A4JQQ7 A0A182M6T9 A0A182GKU6 A0A182WI63 A0A182R9B1 A0A084WN04 A0A182QAQ5 A0A182N7I7 A0A182PEX6 A0A1S4F8X0 D7EJ83 A0A182V5H8 A0A182YJI0 A0A182JYR8 A0A182UDC0 A0A182X244 A0A182FFG9 A0A182T341 A0A182I6A5 Q0IEP6 A0A182J2H7 W5JVR5 A0A182L436 N6UJW0 A0A1W4X4Q9 A0A1W4XEA4 A0A1W4XFH8 A0A1W4XF75 A0A1B0D741 B0WLT2 A0A0C9RVR1 U4TPJ7 W8B4G9 W8BEK3 W8B4G6 A0A034VUZ4 A0A034VXJ3 A0A2A3EFW9 A0A0M3QW65 A0A1B0G7J7 A0A1B0C2P2 A0A034VVX8 A0A034VWM8 V9IEE5 A0A0Q9XAM2 A0A0Q9XMJ2 A0A0Q9XM53 B4KWM5 A0A088ATW5 A0A1I8PXU3 A0A0L0CNP5 A0A0Q9WUM9 A0A1A9ZC18 A0A0Q9WHZ9 A0A1A9XQZ4 A0A1A9UP20 A0A0P8ZWB8 A0A0Q9WSV5 A0A0P8XW46 A0A0P8XW74 A0A0Q9WHH4 A0A0Q9WSZ6 A0A0M9A852 A0A0P8YIX5 A0A1L8DME3 A0A0P9AMA5 A0A0P8YC42 A0A0A1X6B3 A0A1Y1M0Z7 A0A1I8PXW8 A0A0Q9WUJ2 A0A1Y1M549 B3M8W8 X2JAU8-5 A0A026W4P8 B4LHG5 A0A0L7QSK4 A0A0Q5UHK9 A0A154PA73 X2JAU8-2 X2JAU8-6 A0A0Q5U3S5 E0VZW6 A0A0R1E0A2 A0A0Q5UHN1 A0A0R1DUU2 A0A0Q5U2S9 B4QMQ6 A0A0R1E0C3 B4HKL4 X2JAU8 B4N6Z5
PDB
2DL7
E-value=2.02601e-10,
Score=161
Ontologies
GO
GO:0046872
GO:0031594
GO:0055037
GO:0007274
GO:0030833
GO:0044803
GO:2000601
GO:0043325
GO:0045886
GO:0005546
GO:0001881
GO:0042734
GO:0005543
GO:0030424
GO:0008582
GO:0008021
GO:0030054
GO:0005737
GO:0005515
GO:0007264
GO:0007165
GO:0016020
GO:0043565
GO:0043401
GO:0003677
GO:0003707
GO:0007169
GO:0016021
Topology
Subcellular location
Endomembrane system
Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Cell junction Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Synapse Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Cell projection Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Axon Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Presynaptic cell membrane Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Cytoplasmic vesicle Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Secretory vesicle Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Synaptic vesicle Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Recycling endosome Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Cell junction Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Synapse Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Cell projection Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Axon Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Presynaptic cell membrane Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Cytoplasmic vesicle Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Secretory vesicle Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Synaptic vesicle Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
Recycling endosome Detected at presynaptic axon terminals at the neuromuscular junction (PubMed:14980202, PubMed:18701694, PubMed:21464232, PubMed:26686642, PubMed:26567222, PubMed:27601635, PubMed:29568072). Colocalizes with Cdc42 and Rab11 (PubMed:18701694, PubMed:21464232). With evidence from 3 publications.
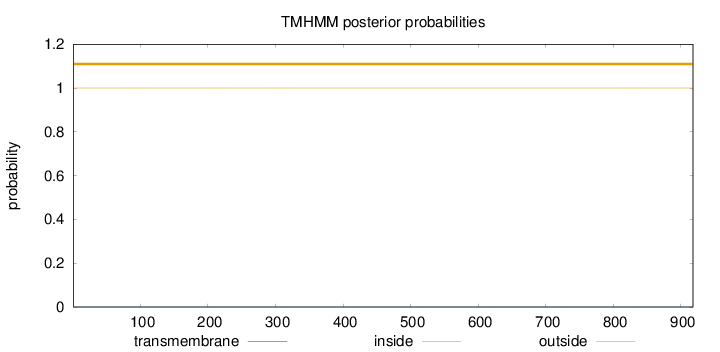
Length:
918
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00018
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00004
outside
1 - 918
Population Genetic Test Statistics
Pi
23.896096
Theta
18.295427
Tajima's D
-1.701202
CLR
0.052267
CSRT
0.0326483675816209
Interpretation
Uncertain
