Pre Gene Modal
BGIBMGA002149
Annotation
PREDICTED:_aarF_domain-containing_protein_kinase_4_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.304
Sequence
CDS
ATGTCACACATTAATGATTTGATAGGTGTAGCCCGTGGCTTACGGCTAGTTGTTGAAGCCAGTACAAAGATGCAGCAGGAAGCATGTAGTATTATGTGGAATAACTCTAGCATTAGGCCACTCCTACAACAATGCCCAACTAACCCACTCACAGCTTATAAACCTAGTGCTGATACTGCAAAAGATGTAATCGAAAGAGCAGTGGTTGTTGCTCATGGCTTAAGGAAATATGTTGTAATGAATGTGCCTAATTTAAACCCTGATATCGAAGGCAAGGTAGAAATGGACCCCAAATTGCAACAGGAGATTGAAGAGTTGAATCGAGAATTCAACAGAACATTTGCCACCTTAGAACAGAAAGTTCAGAATAAAGATAGCCCAGCTGAATTTGTATCACCACTAGAAAATATTAAAATGCAACCTGAGCCATCCAGAATTGAGAAAGATGAAGTTAAAACAGAAATACCCAAGTTTGAAAGTGTTAAACCTAAGTTTGAAACTTTGAGTAATACAACACAATTTGCAACACAAACAATACCAAAACCAGTTGCTAAAAAGAAAATACGTGTATCTCTTAGTGAAAATTCCAAAGCTCGTGTAGTACCATCGTCGAGAATCGGTCGTATGATTTCGTTTGGTTCACTAGCAGCTGGACTCGGAGTCGGCACTATAGCACAGTATGCACGCAATACTTTGCAGTCAGTCACAGGACAAGCCGATGAAGCCGCTAATTCATTTTTGTCTCCAGCTAATGCTGAAAGAATTGTTGACACGCTTTGTAAAGTCAGAGGAGCAGCACTGAAACTAGGTCAATTATTAAGTATACAAGACGAATCTGTGATATCCCCGGAACTTCAACGAATATTCGAGAGAGTACGGCAGTCTGCCGATTTCATGCCAGCATGGCAGGTAGAAAAGGTGATGAGTTCGCAACTTGGACCCGAATGGAGAAGTAAACTACAAAGCTTCGAAGAGAAACCTTTTGCTGCAGCTTCTATAGGGCAAGTGCACTTGGCTGTACTGCATAATGGTCGGGAGGTCGCAATGAAGGTTCAATATCCCGGAGTCGCGAAAGGGATTAACAGTGACATTGACAACCTTGTAGGAGTTATGAAGGTATGGAACATGTTTCCTAAAGGCATGTTCATAGATAACGTTGTGGAAGTGGCGAAAAAAGAACTAGCCTGGGAAGTCGATTATATACGTGAAGCGGAATGTACTAGAAAGTTCAAGACTCTATTGGCACAGTATCCTGAATACTTCGTTCCTGACGTCATAGACGAATTGTGCGCACAGGAAGTGATAACAACAGAACTGATAGACGGCGTACCTCTCGATAAGCTATTCGACGCATCCTATGAGACACGCGTCGATATCGCGTCCAAGATCATGAAGCTGTGCCTCAGAGAAATGTTCGTACTAAGATGTATGCAAACAGATCCGAATTGGGCCAATTTCTTCTACAATACTAATACGAAACAGGTAATATTGTTAGATTTTGGAGCTACAAGAGAGTACTCCAAGGAATTCATGGATCAGTACATTGAGATAATTAAGGCAGCCTCATTGGGCGATCGCGCTGCTATATTGAGAATGTCGCGTGAGATGAAATTCTTGACTGGCTATGAATCTAAAATCATGGAAGAAACGCACGTTGACACTGTCATGATAATGGGCGAAGTATTCACCAGTGAAGGCTCCGGCGATTTCGACTTCGGAACCCAACAAACAACGAGACGGATACAAGCCTTAGTGCCGACCATACTAACGCATAGACTCTGTCCGCCCCCGGAGGAAATATATTCATTGCACAGGAAGCTCTCGGGCGTCTTTCTGCTATGCTCAAAGATGAAAATTAAAATGAACTGCAGGGATATGTTCAATGAGATTTACGATCAATATAAGAGCAACAGCCTCAGGTAA
Protein
MSHINDLIGVARGLRLVVEASTKMQQEACSIMWNNSSIRPLLQQCPTNPLTAYKPSADTAKDVIERAVVVAHGLRKYVVMNVPNLNPDIEGKVEMDPKLQQEIEELNREFNRTFATLEQKVQNKDSPAEFVSPLENIKMQPEPSRIEKDEVKTEIPKFESVKPKFETLSNTTQFATQTIPKPVAKKKIRVSLSENSKARVVPSSRIGRMISFGSLAAGLGVGTIAQYARNTLQSVTGQADEAANSFLSPANAERIVDTLCKVRGAALKLGQLLSIQDESVISPELQRIFERVRQSADFMPAWQVEKVMSSQLGPEWRSKLQSFEEKPFAAASIGQVHLAVLHNGREVAMKVQYPGVAKGINSDIDNLVGVMKVWNMFPKGMFIDNVVEVAKKELAWEVDYIREAECTRKFKTLLAQYPEYFVPDVIDELCAQEVITTELIDGVPLDKLFDASYETRVDIASKIMKLCLREMFVLRCMQTDPNWANFFYNTNTKQVILLDFGATREYSKEFMDQYIEIIKAASLGDRAAILRMSREMKFLTGYESKIMEETHVDTVMIMGEVFTSEGSGDFDFGTQQTTRRIQALVPTILTHRLCPPPEEIYSLHRKLSGVFLLCSKMKIKMNCRDMFNEIYDQYKSNSLR
Summary
Uniprot
H9IY16
A0A2A4JQT9
A0A2H1WYU1
A0A212FHI4
A0A194Q4X4
A0A194QUI2
+ More
A0A0L7LME9 A0A1B6C7G0 A0A232FKA8 K7IV93 A0A1B6ESG4 A0A088ARA7 A0A1B6HZJ3 A0A1B6KC95 A0A1B6I821 A0A1B6IC20 A0A0L7RCX6 A0A0V0G8Q0 E2C531 A0A2A3EDB0 A0A154PIR5 A0A069DW72 A0A0C9RVT1 A0A0K8S8G5 A0A0A9WTR4 A0A146M3J0 E2AZ25 D1ZZY6 A0A0C9QMV2 A0A151XF53 A0A3L8DUP8 A0A2J7Q4Y2 W8B4G4 A0A151I543 A0A067RSN5 A0A158NU38 A0A151J8D4 A0A0C9R1W6 Q29FT4 B4H9S2 F4WUA7 B3MS19 A0A195C7F7 B4Q1Y4 A0A3B0K7T2 A0A1Q3FGP1 A0A1Y1L425 A0A2J7Q4X9 A0A1A9ZEQ6 A0A026X036 A0A0P5UFU6 A0A0P5UFP9 A0A0P5BBL1 A0A0P5ASF7 A0A0N8D045 A0A0N8CHN9 A0A195FK51 B3NX90 A0A0P5WG48 A0A0P5XCA9 A0A0N8D9P3 A0A0P5XNE4 A0A0P5ABQ1 A0A0P4YJ96 A0A0P5WGK4 A0A0N7ZJW3 E9IH46 A0A0P5W7Y3 A0A0P4YXZ6 A0A0P4YWF1 A0A0N8CUM6 A0A0P5B000 A0A0P5W802 A0A0P5AP81 A0A0P5ZMH1 A0A0N7ZQN2 A0A0P5WQF7 B0XC82 A0A1B0ATF8 A0A0L0BTY7 A0A0P5VE69 A0A1I8MKY1 A0A0P5YD38 A0A0P5BTU6 A0A0P5YZF3 A0A1A9XBM6 A0A0P5CF77 A0A0P5WCW9 A0A0P5BKN6 A0A0P5XKJ1 A0A0P5WDC9 A0A0P5WAN8 A0A1J1IDW4 A0A0P5BAJ7 A0A0P5AK48 A0A0P5WD71 A0A034VVK7 A0A034VS47 A0A0P6G5B1
A0A0L7LME9 A0A1B6C7G0 A0A232FKA8 K7IV93 A0A1B6ESG4 A0A088ARA7 A0A1B6HZJ3 A0A1B6KC95 A0A1B6I821 A0A1B6IC20 A0A0L7RCX6 A0A0V0G8Q0 E2C531 A0A2A3EDB0 A0A154PIR5 A0A069DW72 A0A0C9RVT1 A0A0K8S8G5 A0A0A9WTR4 A0A146M3J0 E2AZ25 D1ZZY6 A0A0C9QMV2 A0A151XF53 A0A3L8DUP8 A0A2J7Q4Y2 W8B4G4 A0A151I543 A0A067RSN5 A0A158NU38 A0A151J8D4 A0A0C9R1W6 Q29FT4 B4H9S2 F4WUA7 B3MS19 A0A195C7F7 B4Q1Y4 A0A3B0K7T2 A0A1Q3FGP1 A0A1Y1L425 A0A2J7Q4X9 A0A1A9ZEQ6 A0A026X036 A0A0P5UFU6 A0A0P5UFP9 A0A0P5BBL1 A0A0P5ASF7 A0A0N8D045 A0A0N8CHN9 A0A195FK51 B3NX90 A0A0P5WG48 A0A0P5XCA9 A0A0N8D9P3 A0A0P5XNE4 A0A0P5ABQ1 A0A0P4YJ96 A0A0P5WGK4 A0A0N7ZJW3 E9IH46 A0A0P5W7Y3 A0A0P4YXZ6 A0A0P4YWF1 A0A0N8CUM6 A0A0P5B000 A0A0P5W802 A0A0P5AP81 A0A0P5ZMH1 A0A0N7ZQN2 A0A0P5WQF7 B0XC82 A0A1B0ATF8 A0A0L0BTY7 A0A0P5VE69 A0A1I8MKY1 A0A0P5YD38 A0A0P5BTU6 A0A0P5YZF3 A0A1A9XBM6 A0A0P5CF77 A0A0P5WCW9 A0A0P5BKN6 A0A0P5XKJ1 A0A0P5WDC9 A0A0P5WAN8 A0A1J1IDW4 A0A0P5BAJ7 A0A0P5AK48 A0A0P5WD71 A0A034VVK7 A0A034VS47 A0A0P6G5B1
Pubmed
EMBL
BABH01016007
BABH01016008
BABH01016009
BABH01016010
NWSH01000870
PCG73782.1
+ More
ODYU01012123 SOQ58250.1 AGBW02008503 OWR53203.1 KQ459463 KPJ00592.1 KQ461108 KPJ09127.1 JTDY01000635 KOB76396.1 GEDC01027872 JAS09426.1 NNAY01000096 OXU31023.1 AAZX01001186 GECZ01029126 JAS40643.1 GECU01027626 JAS80080.1 GEBQ01030905 GEBQ01026287 JAT09072.1 JAT13690.1 GECU01024666 JAS83040.1 GECU01023202 JAS84504.1 KQ414615 KOC68611.1 GECL01001723 JAP04401.1 GL452737 EFN76939.1 KZ288289 PBC29282.1 KQ434931 KZC11761.1 GBGD01000798 JAC88091.1 GBYB01001988 GBYB01001989 GBYB01011821 JAG71755.1 JAG71756.1 JAG81588.1 GBRD01016362 JAG49464.1 GBHO01031732 JAG11872.1 GDHC01004790 GDHC01004281 JAQ13839.1 JAQ14348.1 GL444070 EFN61320.1 KQ971338 EFA01775.2 GBYB01001987 JAG71754.1 KQ982211 KYQ58965.1 QOIP01000004 RLU23468.1 NEVH01018373 PNF23634.1 GAMC01010540 JAB96015.1 KQ976445 KYM86114.1 KK852484 KDR22829.1 ADTU01026190 KQ979585 KYN20633.1 GBYB01001990 JAG71757.1 CH379065 EAL31495.1 CH479231 EDW36569.1 GL888355 EGI62215.1 CH902622 EDV34574.1 KQ978143 KYM96809.1 CM000162 EDX01505.1 OUUW01000021 SPP89383.1 GFDL01008340 JAV26705.1 GEZM01068913 GEZM01068912 JAV66705.1 PNF23635.1 KK107054 EZA61612.1 GDIP01113650 JAL90064.1 GDIP01113649 JAL90065.1 GDIP01202227 JAJ21175.1 GDIP01225723 GDIP01218426 GDIP01216476 GDIP01211581 GDIP01196277 GDIP01195133 GDIP01195131 GDIP01149799 GDIP01132145 GDIP01130737 GDIP01130736 GDIP01126535 GDIP01125244 GDIP01112398 GDIP01107195 GDIP01104537 GDIP01046249 JAJ28269.1 GDIP01113651 GDIP01081887 JAM21828.1 GDIP01196276 GDIP01195134 GDIP01149797 GDIP01132143 GDIP01130735 GDIP01126536 GDIP01125243 GDIP01124190 JAL72979.1 KQ981522 KYN40731.1 CH954180 EDV47262.1 GDIP01086596 JAM17119.1 GDIP01236876 GDIP01236875 GDIP01097240 GDIP01074202 GDIP01061393 GDIP01055105 GDIP01052302 JAM29513.1 GDIP01097238 GDIP01055103 JAM48612.1 GDIP01147195 GDIP01074201 GDIP01071955 GDIP01044256 LRGB01000915 JAM31760.1 KZS15173.1 GDIP01202229 GDIP01202228 GDIP01202226 JAJ21174.1 GDIP01238234 GDIP01229880 GDIP01229879 GDIP01227980 GDIP01208079 GDIP01208078 GDIP01132144 GDIP01126537 GDIP01096082 GDIP01085348 GDIP01085347 GDIP01046248 JAI95421.1 GDIP01089954 GDIP01086598 GDIP01086597 JAM17118.1 GDIP01238233 GDIP01096081 JAI85168.1 GL763174 EFZ20095.1 GDIP01201083 GDIP01201082 GDIP01089955 JAM13760.1 GDIP01220984 GDIP01220982 GDIP01137530 GDIP01062848 JAJ02420.1 GDIP01222083 GDIP01222082 JAJ01320.1 GDIP01193676 GDIP01145689 GDIP01097239 GDIP01074203 GDIP01071957 GDIP01071956 GDIP01044258 GDIP01044257 GDIP01007131 JAM06476.1 GDIP01220983 GDIP01191883 GDIP01167329 GDIP01164836 GDIP01149798 GDIP01140832 GDIP01137531 GDIP01062849 GDIP01062847 JAJ31519.1 GDIP01201081 GDIP01089953 JAM13762.1 GDIP01199726 JAJ23676.1 GDIP01055104 JAM48611.1 GDIP01231229 GDIP01222081 GDIP01123083 JAJ01321.1 GDIP01199773 GDIP01199772 GDIP01168984 GDIP01083796 JAM19919.1 DS232677 EDS44688.1 JXJN01003292 JRES01001343 KNC23545.1 GDIP01101593 JAM02122.1 GDIP01059396 JAM44319.1 GDIP01195132 GDIP01051100 JAJ28270.1 GDIP01195135 GDIP01051101 JAM52614.1 GDIP01187178 JAJ36224.1 GDIP01088788 JAM14927.1 GDIP01198186 JAJ25216.1 GDIP01072942 JAM30773.1 GDIP01087692 JAM16023.1 GDIP01088790 GDIP01088789 JAM14926.1 CVRI01000047 CRK97190.1 GDIP01187179 JAJ36223.1 GDIP01198188 GDIP01198187 JAJ25214.1 GDIP01087694 GDIP01087693 JAM16022.1 GAKP01012825 JAC46127.1 GAKP01012826 JAC46126.1 GDIQ01056898 JAN37839.1
ODYU01012123 SOQ58250.1 AGBW02008503 OWR53203.1 KQ459463 KPJ00592.1 KQ461108 KPJ09127.1 JTDY01000635 KOB76396.1 GEDC01027872 JAS09426.1 NNAY01000096 OXU31023.1 AAZX01001186 GECZ01029126 JAS40643.1 GECU01027626 JAS80080.1 GEBQ01030905 GEBQ01026287 JAT09072.1 JAT13690.1 GECU01024666 JAS83040.1 GECU01023202 JAS84504.1 KQ414615 KOC68611.1 GECL01001723 JAP04401.1 GL452737 EFN76939.1 KZ288289 PBC29282.1 KQ434931 KZC11761.1 GBGD01000798 JAC88091.1 GBYB01001988 GBYB01001989 GBYB01011821 JAG71755.1 JAG71756.1 JAG81588.1 GBRD01016362 JAG49464.1 GBHO01031732 JAG11872.1 GDHC01004790 GDHC01004281 JAQ13839.1 JAQ14348.1 GL444070 EFN61320.1 KQ971338 EFA01775.2 GBYB01001987 JAG71754.1 KQ982211 KYQ58965.1 QOIP01000004 RLU23468.1 NEVH01018373 PNF23634.1 GAMC01010540 JAB96015.1 KQ976445 KYM86114.1 KK852484 KDR22829.1 ADTU01026190 KQ979585 KYN20633.1 GBYB01001990 JAG71757.1 CH379065 EAL31495.1 CH479231 EDW36569.1 GL888355 EGI62215.1 CH902622 EDV34574.1 KQ978143 KYM96809.1 CM000162 EDX01505.1 OUUW01000021 SPP89383.1 GFDL01008340 JAV26705.1 GEZM01068913 GEZM01068912 JAV66705.1 PNF23635.1 KK107054 EZA61612.1 GDIP01113650 JAL90064.1 GDIP01113649 JAL90065.1 GDIP01202227 JAJ21175.1 GDIP01225723 GDIP01218426 GDIP01216476 GDIP01211581 GDIP01196277 GDIP01195133 GDIP01195131 GDIP01149799 GDIP01132145 GDIP01130737 GDIP01130736 GDIP01126535 GDIP01125244 GDIP01112398 GDIP01107195 GDIP01104537 GDIP01046249 JAJ28269.1 GDIP01113651 GDIP01081887 JAM21828.1 GDIP01196276 GDIP01195134 GDIP01149797 GDIP01132143 GDIP01130735 GDIP01126536 GDIP01125243 GDIP01124190 JAL72979.1 KQ981522 KYN40731.1 CH954180 EDV47262.1 GDIP01086596 JAM17119.1 GDIP01236876 GDIP01236875 GDIP01097240 GDIP01074202 GDIP01061393 GDIP01055105 GDIP01052302 JAM29513.1 GDIP01097238 GDIP01055103 JAM48612.1 GDIP01147195 GDIP01074201 GDIP01071955 GDIP01044256 LRGB01000915 JAM31760.1 KZS15173.1 GDIP01202229 GDIP01202228 GDIP01202226 JAJ21174.1 GDIP01238234 GDIP01229880 GDIP01229879 GDIP01227980 GDIP01208079 GDIP01208078 GDIP01132144 GDIP01126537 GDIP01096082 GDIP01085348 GDIP01085347 GDIP01046248 JAI95421.1 GDIP01089954 GDIP01086598 GDIP01086597 JAM17118.1 GDIP01238233 GDIP01096081 JAI85168.1 GL763174 EFZ20095.1 GDIP01201083 GDIP01201082 GDIP01089955 JAM13760.1 GDIP01220984 GDIP01220982 GDIP01137530 GDIP01062848 JAJ02420.1 GDIP01222083 GDIP01222082 JAJ01320.1 GDIP01193676 GDIP01145689 GDIP01097239 GDIP01074203 GDIP01071957 GDIP01071956 GDIP01044258 GDIP01044257 GDIP01007131 JAM06476.1 GDIP01220983 GDIP01191883 GDIP01167329 GDIP01164836 GDIP01149798 GDIP01140832 GDIP01137531 GDIP01062849 GDIP01062847 JAJ31519.1 GDIP01201081 GDIP01089953 JAM13762.1 GDIP01199726 JAJ23676.1 GDIP01055104 JAM48611.1 GDIP01231229 GDIP01222081 GDIP01123083 JAJ01321.1 GDIP01199773 GDIP01199772 GDIP01168984 GDIP01083796 JAM19919.1 DS232677 EDS44688.1 JXJN01003292 JRES01001343 KNC23545.1 GDIP01101593 JAM02122.1 GDIP01059396 JAM44319.1 GDIP01195132 GDIP01051100 JAJ28270.1 GDIP01195135 GDIP01051101 JAM52614.1 GDIP01187178 JAJ36224.1 GDIP01088788 JAM14927.1 GDIP01198186 JAJ25216.1 GDIP01072942 JAM30773.1 GDIP01087692 JAM16023.1 GDIP01088790 GDIP01088789 JAM14926.1 CVRI01000047 CRK97190.1 GDIP01187179 JAJ36223.1 GDIP01198188 GDIP01198187 JAJ25214.1 GDIP01087694 GDIP01087693 JAM16022.1 GAKP01012825 JAC46127.1 GAKP01012826 JAC46126.1 GDIQ01056898 JAN37839.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000215335 UP000002358 UP000005203 UP000053825 UP000008237 UP000242457 UP000076502 UP000000311 UP000007266 UP000075809 UP000279307 UP000235965 UP000078540 UP000027135 UP000005205 UP000078492 UP000001819 UP000008744 UP000007755 UP000007801 UP000078542 UP000002282 UP000268350 UP000092445 UP000053097 UP000078541 UP000008711 UP000076858 UP000002320 UP000092460 UP000037069 UP000095301 UP000092443 UP000183832
UP000215335 UP000002358 UP000005203 UP000053825 UP000008237 UP000242457 UP000076502 UP000000311 UP000007266 UP000075809 UP000279307 UP000235965 UP000078540 UP000027135 UP000005205 UP000078492 UP000001819 UP000008744 UP000007755 UP000007801 UP000078542 UP000002282 UP000268350 UP000092445 UP000053097 UP000078541 UP000008711 UP000076858 UP000002320 UP000092460 UP000037069 UP000095301 UP000092443 UP000183832
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IY16
A0A2A4JQT9
A0A2H1WYU1
A0A212FHI4
A0A194Q4X4
A0A194QUI2
+ More
A0A0L7LME9 A0A1B6C7G0 A0A232FKA8 K7IV93 A0A1B6ESG4 A0A088ARA7 A0A1B6HZJ3 A0A1B6KC95 A0A1B6I821 A0A1B6IC20 A0A0L7RCX6 A0A0V0G8Q0 E2C531 A0A2A3EDB0 A0A154PIR5 A0A069DW72 A0A0C9RVT1 A0A0K8S8G5 A0A0A9WTR4 A0A146M3J0 E2AZ25 D1ZZY6 A0A0C9QMV2 A0A151XF53 A0A3L8DUP8 A0A2J7Q4Y2 W8B4G4 A0A151I543 A0A067RSN5 A0A158NU38 A0A151J8D4 A0A0C9R1W6 Q29FT4 B4H9S2 F4WUA7 B3MS19 A0A195C7F7 B4Q1Y4 A0A3B0K7T2 A0A1Q3FGP1 A0A1Y1L425 A0A2J7Q4X9 A0A1A9ZEQ6 A0A026X036 A0A0P5UFU6 A0A0P5UFP9 A0A0P5BBL1 A0A0P5ASF7 A0A0N8D045 A0A0N8CHN9 A0A195FK51 B3NX90 A0A0P5WG48 A0A0P5XCA9 A0A0N8D9P3 A0A0P5XNE4 A0A0P5ABQ1 A0A0P4YJ96 A0A0P5WGK4 A0A0N7ZJW3 E9IH46 A0A0P5W7Y3 A0A0P4YXZ6 A0A0P4YWF1 A0A0N8CUM6 A0A0P5B000 A0A0P5W802 A0A0P5AP81 A0A0P5ZMH1 A0A0N7ZQN2 A0A0P5WQF7 B0XC82 A0A1B0ATF8 A0A0L0BTY7 A0A0P5VE69 A0A1I8MKY1 A0A0P5YD38 A0A0P5BTU6 A0A0P5YZF3 A0A1A9XBM6 A0A0P5CF77 A0A0P5WCW9 A0A0P5BKN6 A0A0P5XKJ1 A0A0P5WDC9 A0A0P5WAN8 A0A1J1IDW4 A0A0P5BAJ7 A0A0P5AK48 A0A0P5WD71 A0A034VVK7 A0A034VS47 A0A0P6G5B1
A0A0L7LME9 A0A1B6C7G0 A0A232FKA8 K7IV93 A0A1B6ESG4 A0A088ARA7 A0A1B6HZJ3 A0A1B6KC95 A0A1B6I821 A0A1B6IC20 A0A0L7RCX6 A0A0V0G8Q0 E2C531 A0A2A3EDB0 A0A154PIR5 A0A069DW72 A0A0C9RVT1 A0A0K8S8G5 A0A0A9WTR4 A0A146M3J0 E2AZ25 D1ZZY6 A0A0C9QMV2 A0A151XF53 A0A3L8DUP8 A0A2J7Q4Y2 W8B4G4 A0A151I543 A0A067RSN5 A0A158NU38 A0A151J8D4 A0A0C9R1W6 Q29FT4 B4H9S2 F4WUA7 B3MS19 A0A195C7F7 B4Q1Y4 A0A3B0K7T2 A0A1Q3FGP1 A0A1Y1L425 A0A2J7Q4X9 A0A1A9ZEQ6 A0A026X036 A0A0P5UFU6 A0A0P5UFP9 A0A0P5BBL1 A0A0P5ASF7 A0A0N8D045 A0A0N8CHN9 A0A195FK51 B3NX90 A0A0P5WG48 A0A0P5XCA9 A0A0N8D9P3 A0A0P5XNE4 A0A0P5ABQ1 A0A0P4YJ96 A0A0P5WGK4 A0A0N7ZJW3 E9IH46 A0A0P5W7Y3 A0A0P4YXZ6 A0A0P4YWF1 A0A0N8CUM6 A0A0P5B000 A0A0P5W802 A0A0P5AP81 A0A0P5ZMH1 A0A0N7ZQN2 A0A0P5WQF7 B0XC82 A0A1B0ATF8 A0A0L0BTY7 A0A0P5VE69 A0A1I8MKY1 A0A0P5YD38 A0A0P5BTU6 A0A0P5YZF3 A0A1A9XBM6 A0A0P5CF77 A0A0P5WCW9 A0A0P5BKN6 A0A0P5XKJ1 A0A0P5WDC9 A0A0P5WAN8 A0A1J1IDW4 A0A0P5BAJ7 A0A0P5AK48 A0A0P5WD71 A0A034VVK7 A0A034VS47 A0A0P6G5B1
PDB
4PED
E-value=1.60073e-136,
Score=1247
Ontologies
KEGG
GO
PANTHER
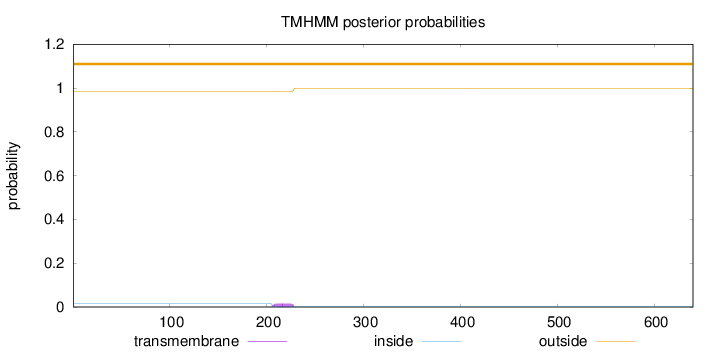
Topology
Length:
640
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.345130000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01562
outside
1 - 640
Population Genetic Test Statistics
Pi
1.088715
Theta
5.300244
Tajima's D
-2.54524
CLR
4.737339
CSRT
0.000349982500874956
Interpretation
Possibly Positive selection
