Gene
KWMTBOMO00153
Pre Gene Modal
BGIBMGA001998
Annotation
PREDICTED:_protein_GPR107_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.768
Sequence
CDS
ATGTTTCGCGTAATGAAAACAACTTTCCTGTACATCCTGATGGTTGTGGCTTCAGTTTCATGCAGAATACACAAACTGCTTCTTACGGATGATCAACGGAAGTATGTGGACATCACGACTTTCGGATTCCTTGAGCGTGGTATCCTAGAAGTCAATTTAGTTAACTTTATGATGCCTTCAGGACCTTTCGGGGAGTACGGCTTCACATTGGACCGAACAATAAATGACGCGATTAGTCCGTATTTAGCTCAAGGAAATTCCAATTCATGTTTTCTGAAAGAAAATGTAATTACTCCACAACACCCAGCCATAGTTTTCCTTAAAATGGATTTCGAAAACTTAAGGTTAAACGTTACATGCCGAGCGGCAAGCCAGCATATATATAGAAACAAAAGCGTCATTCCAAGAGTTTTCCCTAATGATGCAGAAGAAAAATGTTCATATGATTTGCTACCTATTCAAAGGGAAACCAGAAATCAAATAGACTACTATAATACAAGTTTTGCTCTGTACATAACCTCTAAGAAGGAAGAAGGTCTGTACAACTTGTACTTCAATAACTGTCCCTATAAAGGCGAAGGGAACCCGACACCTGTTAATGCAATAATTGAAATCTTCGAGAGCAACGAAGGAAACTACCTGTCCGCTGGTGAGATCCCGCTGCCTCTATTATATGCAGTGACGTCATCGATATTCTTGTTCTGCGCTATATTGTGGACGTACATTCTGAAGACGAGCAGACAGACCGTGTTCCGCATACACATAATAATGTGCATCCTGGTGTATCTGAAGGCAGCCTCGCTTGCCTTCCACGGGATAAACTATCACTTCATACAGATAAGGGGCGAGCACGTGACGGCCTGGGCGATATTGTATTATATCACCCATCTGCTTAAAGGGGCAGTTCTGTTCGTGACTATAGTTCTTGTCGGCACCGGATGGAATTTCATCAAGCATATACTGTCCGATCGCGATAAGAAACTGTTCATGATTGTTATACCATTGCAGGTGCTAGCTAATGTTGCAGAGATAATATTAGCGGAAAGCGAGGAAGGCGCAGTCGAACACAACGCCTGGCGCGATATCTTTATAGTTGTCGAGCTAGTTTGTTGCGTCGCTATTGTTTTCCCTGTTGTCTGGTCGATACGACATTTACAAGACGCATCGACGACTGACGGCAAAGCGGCCATCAATCTTCGCAAACTGAAACTATTCCAGCATTTCTACGTGATGGTCGCCTGTTACATATACTTTACGAGGATTGTGGTCTTCATATTGAAGAAAACAGTGCGGTTCCAATACTCGTGGATGGACGAGATGTTCCGCGAGCTGGCCACACTCGTGTTCTTCCTGATGACAGGCTACAAGTTCAGGCCGGCGGCCGCTAACCCGTACTTCACGCCGACCCACTTCGATGACGTCAAACCCGAAGTATTATCGGATATCGGTATACTGGAAGGCGTGACACGAATACCGAATCGGGGCGAGAGAAGACGGGAAGACCTCCAGCCTTTAATGCAGGACGCTGACTACAACTCTGATTAA
Protein
MFRVMKTTFLYILMVVASVSCRIHKLLLTDDQRKYVDITTFGFLERGILEVNLVNFMMPSGPFGEYGFTLDRTINDAISPYLAQGNSNSCFLKENVITPQHPAIVFLKMDFENLRLNVTCRAASQHIYRNKSVIPRVFPNDAEEKCSYDLLPIQRETRNQIDYYNTSFALYITSKKEEGLYNLYFNNCPYKGEGNPTPVNAIIEIFESNEGNYLSAGEIPLPLLYAVTSSIFLFCAILWTYILKTSRQTVFRIHIIMCILVYLKAASLAFHGINYHFIQIRGEHVTAWAILYYITHLLKGAVLFVTIVLVGTGWNFIKHILSDRDKKLFMIVIPLQVLANVAEIILAESEEGAVEHNAWRDIFIVVELVCCVAIVFPVVWSIRHLQDASTTDGKAAINLRKLKLFQHFYVMVACYIYFTRIVVFILKKTVRFQYSWMDEMFRELATLVFFLMTGYKFRPAAANPYFTPTHFDDVKPEVLSDIGILEGVTRIPNRGERRREDLQPLMQDADYNSD
Summary
Uniprot
A0A2A4JQU4
A0A1E1WCL5
A0A212FHK6
A0A194QVY3
D6WH46
A0A084VVB2
+ More
A0A1B6HIZ1 A0A1B6FL11 A0A1B6FMA9 A0A182IT87 A0A1B6L602 A0A1L8DJW4 A0A1L8DK02 A0A182RKC6 A0A023EUZ3 A0A336LRC1 A0A1B6CWU3 U5ESJ2 A0A1S4G7U1 A0A182JXB8 A0A2J7RSE4 A0A023F2C1 A0A224XM90 A0A1Q3FYB4 A0A1I8JT72 A0A182XNK0 A0A0L7QS86 A0A182U7L8 A0A182YH85 Q7Q4R4 A0A182VP60 A0A182MWI8 A0A182VT21 A0A2M3Z5M5 A0A182QWL3 A0A182PL29 A0A232EUQ1 A0A2M4AK96 W5JKB8 A0A2M4AK12 K7JHR5 A0A0A9VZP7 A0A182FIW7 A0A2M4BIE6 A0A2M4BHT2 A0A182MZP6 A0A0C9S0P6 A0A0C9RLD8 A0A0M9A6V7 A0A088A3L3 A0A195F1T4 E2AKS8 A0A2A3ESK2 A0A1Y1KZP0 A0A026WSB0 A0A0J7NH29 A0A158NL14 A0A1J1IPI6 E2BVA5 J3JXG1 N6T081 A0A1W6EVT9 A0A1B0CME1 A0A195BXS5 A0A195BHE4 F4WG08 E9IN22 A0A195EFL9 A0A151XBP7 A0A1Q3FHF0 A0A2P8Z9Y0 B0XFM1 A0A3Q0IMY4 A0A1E1X4V5 A0A023GG65 A0A2S2NEG6 A0A2S2QUK5 A0A2S2RB53 V5I9W8 A0A023FQ43 A0A2H8TZH7 A0A224Y3I7 J9K7U0 A0A164X9F0 A0A0P5K1A6 A0A0N7ZXY2 A0A0P5WFL9 A0A0C9Q2V2 A0A0P5TY56 A0A0P5ZC73 A0A0P5LQ75 A0A0P6B4X8 A0A0P6CSR3 A0A0P6DFV0
A0A1B6HIZ1 A0A1B6FL11 A0A1B6FMA9 A0A182IT87 A0A1B6L602 A0A1L8DJW4 A0A1L8DK02 A0A182RKC6 A0A023EUZ3 A0A336LRC1 A0A1B6CWU3 U5ESJ2 A0A1S4G7U1 A0A182JXB8 A0A2J7RSE4 A0A023F2C1 A0A224XM90 A0A1Q3FYB4 A0A1I8JT72 A0A182XNK0 A0A0L7QS86 A0A182U7L8 A0A182YH85 Q7Q4R4 A0A182VP60 A0A182MWI8 A0A182VT21 A0A2M3Z5M5 A0A182QWL3 A0A182PL29 A0A232EUQ1 A0A2M4AK96 W5JKB8 A0A2M4AK12 K7JHR5 A0A0A9VZP7 A0A182FIW7 A0A2M4BIE6 A0A2M4BHT2 A0A182MZP6 A0A0C9S0P6 A0A0C9RLD8 A0A0M9A6V7 A0A088A3L3 A0A195F1T4 E2AKS8 A0A2A3ESK2 A0A1Y1KZP0 A0A026WSB0 A0A0J7NH29 A0A158NL14 A0A1J1IPI6 E2BVA5 J3JXG1 N6T081 A0A1W6EVT9 A0A1B0CME1 A0A195BXS5 A0A195BHE4 F4WG08 E9IN22 A0A195EFL9 A0A151XBP7 A0A1Q3FHF0 A0A2P8Z9Y0 B0XFM1 A0A3Q0IMY4 A0A1E1X4V5 A0A023GG65 A0A2S2NEG6 A0A2S2QUK5 A0A2S2RB53 V5I9W8 A0A023FQ43 A0A2H8TZH7 A0A224Y3I7 J9K7U0 A0A164X9F0 A0A0P5K1A6 A0A0N7ZXY2 A0A0P5WFL9 A0A0C9Q2V2 A0A0P5TY56 A0A0P5ZC73 A0A0P5LQ75 A0A0P6B4X8 A0A0P6CSR3 A0A0P6DFV0
Pubmed
EMBL
NWSH01000870
PCG73780.1
GDQN01006355
JAT84699.1
AGBW02008503
OWR53204.1
+ More
KQ461108 KPJ09125.1 KQ971330 EEZ99755.1 ATLV01017170 KE525157 KFB41906.1 GECU01033049 JAS74657.1 GECZ01018900 JAS50869.1 GECZ01018468 JAS51301.1 GEBQ01020829 JAT19148.1 GFDF01007332 JAV06752.1 GFDF01007333 JAV06751.1 GAPW01000786 JAC12812.1 UFQT01000128 SSX20574.1 GEDC01019414 JAS17884.1 GANO01002330 JAB57541.1 NEVH01000261 PNF43748.1 GBBI01003568 JAC15144.1 GFTR01007173 JAW09253.1 GFDL01002470 JAV32575.1 APCN01005721 APCN01005722 KQ414758 KOC61502.1 AAAB01008963 EAA12050.5 AXCM01002513 GGFM01003076 MBW23827.1 AXCN02002488 NNAY01002105 OXU22073.1 GGFK01007717 MBW41038.1 ADMH02000958 ETN64571.1 GGFK01007751 MBW41072.1 GBHO01042989 GBHO01042988 GBRD01005185 GDHC01008598 GDHC01007947 JAG00615.1 JAG00616.1 JAG60636.1 JAQ10031.1 JAQ10682.1 GGFJ01003643 MBW52784.1 GGFJ01003475 MBW52616.1 GBYB01014161 JAG83928.1 GBYB01014162 JAG83929.1 KQ435724 KOX78450.1 KQ981866 KYN34132.1 GL440405 EFN65951.1 KZ288187 PBC34708.1 GEZM01071150 JAV66008.1 KK107111 QOIP01000011 EZA58935.1 RLU16445.1 LBMM01005080 KMQ91830.1 ADTU01019140 CVRI01000054 CRL00409.1 GL450824 EFN80397.1 BT127930 AEE62892.1 APGK01057456 KB741280 KB632033 ENN70918.1 ERL88207.1 KY563429 ARK19838.1 AJWK01018611 KQ978501 KYM93459.1 KQ976488 KYM83574.1 GL888128 EGI66775.1 GL764272 EFZ18027.1 KQ978957 KYN26961.1 KQ982320 KYQ57783.1 GFDL01008067 JAV26978.1 PYGN01000132 PSN53305.1 DS232940 EDS26884.1 GFAC01004903 JAT94285.1 GBBM01003440 JAC31978.1 GGMR01003002 MBY15621.1 GGMS01012215 MBY81418.1 GGMS01017369 MBY86572.1 GALX01002210 JAB66256.1 GBBK01001679 JAC22803.1 GFXV01007842 MBW19647.1 GFPF01000981 MAA12127.1 ABLF02029285 LRGB01001005 KZS13991.1 GDIQ01190610 JAK61115.1 GDIP01201681 JAJ21721.1 GDIP01087499 JAM16216.1 GBYB01008363 JAG78130.1 GDIP01124901 JAL78813.1 GDIP01046010 JAM57705.1 GDIQ01175207 JAK76518.1 GDIP01022057 JAM81658.1 GDIQ01120533 GDIQ01087845 GDIQ01077995 GDIQ01030274 JAN06892.1 GDIQ01120531 GDIQ01081594 GDIQ01077994 JAN16743.1
KQ461108 KPJ09125.1 KQ971330 EEZ99755.1 ATLV01017170 KE525157 KFB41906.1 GECU01033049 JAS74657.1 GECZ01018900 JAS50869.1 GECZ01018468 JAS51301.1 GEBQ01020829 JAT19148.1 GFDF01007332 JAV06752.1 GFDF01007333 JAV06751.1 GAPW01000786 JAC12812.1 UFQT01000128 SSX20574.1 GEDC01019414 JAS17884.1 GANO01002330 JAB57541.1 NEVH01000261 PNF43748.1 GBBI01003568 JAC15144.1 GFTR01007173 JAW09253.1 GFDL01002470 JAV32575.1 APCN01005721 APCN01005722 KQ414758 KOC61502.1 AAAB01008963 EAA12050.5 AXCM01002513 GGFM01003076 MBW23827.1 AXCN02002488 NNAY01002105 OXU22073.1 GGFK01007717 MBW41038.1 ADMH02000958 ETN64571.1 GGFK01007751 MBW41072.1 GBHO01042989 GBHO01042988 GBRD01005185 GDHC01008598 GDHC01007947 JAG00615.1 JAG00616.1 JAG60636.1 JAQ10031.1 JAQ10682.1 GGFJ01003643 MBW52784.1 GGFJ01003475 MBW52616.1 GBYB01014161 JAG83928.1 GBYB01014162 JAG83929.1 KQ435724 KOX78450.1 KQ981866 KYN34132.1 GL440405 EFN65951.1 KZ288187 PBC34708.1 GEZM01071150 JAV66008.1 KK107111 QOIP01000011 EZA58935.1 RLU16445.1 LBMM01005080 KMQ91830.1 ADTU01019140 CVRI01000054 CRL00409.1 GL450824 EFN80397.1 BT127930 AEE62892.1 APGK01057456 KB741280 KB632033 ENN70918.1 ERL88207.1 KY563429 ARK19838.1 AJWK01018611 KQ978501 KYM93459.1 KQ976488 KYM83574.1 GL888128 EGI66775.1 GL764272 EFZ18027.1 KQ978957 KYN26961.1 KQ982320 KYQ57783.1 GFDL01008067 JAV26978.1 PYGN01000132 PSN53305.1 DS232940 EDS26884.1 GFAC01004903 JAT94285.1 GBBM01003440 JAC31978.1 GGMR01003002 MBY15621.1 GGMS01012215 MBY81418.1 GGMS01017369 MBY86572.1 GALX01002210 JAB66256.1 GBBK01001679 JAC22803.1 GFXV01007842 MBW19647.1 GFPF01000981 MAA12127.1 ABLF02029285 LRGB01001005 KZS13991.1 GDIQ01190610 JAK61115.1 GDIP01201681 JAJ21721.1 GDIP01087499 JAM16216.1 GBYB01008363 JAG78130.1 GDIP01124901 JAL78813.1 GDIP01046010 JAM57705.1 GDIQ01175207 JAK76518.1 GDIP01022057 JAM81658.1 GDIQ01120533 GDIQ01087845 GDIQ01077995 GDIQ01030274 JAN06892.1 GDIQ01120531 GDIQ01081594 GDIQ01077994 JAN16743.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000007266
UP000030765
UP000075880
+ More
UP000075900 UP000075881 UP000235965 UP000075840 UP000076407 UP000053825 UP000075902 UP000076408 UP000007062 UP000075903 UP000075883 UP000075920 UP000075886 UP000075885 UP000215335 UP000000673 UP000002358 UP000069272 UP000075884 UP000053105 UP000005203 UP000078541 UP000000311 UP000242457 UP000053097 UP000279307 UP000036403 UP000005205 UP000183832 UP000008237 UP000019118 UP000030742 UP000092461 UP000078542 UP000078540 UP000007755 UP000078492 UP000075809 UP000245037 UP000002320 UP000079169 UP000007819 UP000076858
UP000075900 UP000075881 UP000235965 UP000075840 UP000076407 UP000053825 UP000075902 UP000076408 UP000007062 UP000075903 UP000075883 UP000075920 UP000075886 UP000075885 UP000215335 UP000000673 UP000002358 UP000069272 UP000075884 UP000053105 UP000005203 UP000078541 UP000000311 UP000242457 UP000053097 UP000279307 UP000036403 UP000005205 UP000183832 UP000008237 UP000019118 UP000030742 UP000092461 UP000078542 UP000078540 UP000007755 UP000078492 UP000075809 UP000245037 UP000002320 UP000079169 UP000007819 UP000076858
Pfam
PF06814 Lung_7-TM_R
Interpro
IPR009637
GPR107-like
ProteinModelPortal
A0A2A4JQU4
A0A1E1WCL5
A0A212FHK6
A0A194QVY3
D6WH46
A0A084VVB2
+ More
A0A1B6HIZ1 A0A1B6FL11 A0A1B6FMA9 A0A182IT87 A0A1B6L602 A0A1L8DJW4 A0A1L8DK02 A0A182RKC6 A0A023EUZ3 A0A336LRC1 A0A1B6CWU3 U5ESJ2 A0A1S4G7U1 A0A182JXB8 A0A2J7RSE4 A0A023F2C1 A0A224XM90 A0A1Q3FYB4 A0A1I8JT72 A0A182XNK0 A0A0L7QS86 A0A182U7L8 A0A182YH85 Q7Q4R4 A0A182VP60 A0A182MWI8 A0A182VT21 A0A2M3Z5M5 A0A182QWL3 A0A182PL29 A0A232EUQ1 A0A2M4AK96 W5JKB8 A0A2M4AK12 K7JHR5 A0A0A9VZP7 A0A182FIW7 A0A2M4BIE6 A0A2M4BHT2 A0A182MZP6 A0A0C9S0P6 A0A0C9RLD8 A0A0M9A6V7 A0A088A3L3 A0A195F1T4 E2AKS8 A0A2A3ESK2 A0A1Y1KZP0 A0A026WSB0 A0A0J7NH29 A0A158NL14 A0A1J1IPI6 E2BVA5 J3JXG1 N6T081 A0A1W6EVT9 A0A1B0CME1 A0A195BXS5 A0A195BHE4 F4WG08 E9IN22 A0A195EFL9 A0A151XBP7 A0A1Q3FHF0 A0A2P8Z9Y0 B0XFM1 A0A3Q0IMY4 A0A1E1X4V5 A0A023GG65 A0A2S2NEG6 A0A2S2QUK5 A0A2S2RB53 V5I9W8 A0A023FQ43 A0A2H8TZH7 A0A224Y3I7 J9K7U0 A0A164X9F0 A0A0P5K1A6 A0A0N7ZXY2 A0A0P5WFL9 A0A0C9Q2V2 A0A0P5TY56 A0A0P5ZC73 A0A0P5LQ75 A0A0P6B4X8 A0A0P6CSR3 A0A0P6DFV0
A0A1B6HIZ1 A0A1B6FL11 A0A1B6FMA9 A0A182IT87 A0A1B6L602 A0A1L8DJW4 A0A1L8DK02 A0A182RKC6 A0A023EUZ3 A0A336LRC1 A0A1B6CWU3 U5ESJ2 A0A1S4G7U1 A0A182JXB8 A0A2J7RSE4 A0A023F2C1 A0A224XM90 A0A1Q3FYB4 A0A1I8JT72 A0A182XNK0 A0A0L7QS86 A0A182U7L8 A0A182YH85 Q7Q4R4 A0A182VP60 A0A182MWI8 A0A182VT21 A0A2M3Z5M5 A0A182QWL3 A0A182PL29 A0A232EUQ1 A0A2M4AK96 W5JKB8 A0A2M4AK12 K7JHR5 A0A0A9VZP7 A0A182FIW7 A0A2M4BIE6 A0A2M4BHT2 A0A182MZP6 A0A0C9S0P6 A0A0C9RLD8 A0A0M9A6V7 A0A088A3L3 A0A195F1T4 E2AKS8 A0A2A3ESK2 A0A1Y1KZP0 A0A026WSB0 A0A0J7NH29 A0A158NL14 A0A1J1IPI6 E2BVA5 J3JXG1 N6T081 A0A1W6EVT9 A0A1B0CME1 A0A195BXS5 A0A195BHE4 F4WG08 E9IN22 A0A195EFL9 A0A151XBP7 A0A1Q3FHF0 A0A2P8Z9Y0 B0XFM1 A0A3Q0IMY4 A0A1E1X4V5 A0A023GG65 A0A2S2NEG6 A0A2S2QUK5 A0A2S2RB53 V5I9W8 A0A023FQ43 A0A2H8TZH7 A0A224Y3I7 J9K7U0 A0A164X9F0 A0A0P5K1A6 A0A0N7ZXY2 A0A0P5WFL9 A0A0C9Q2V2 A0A0P5TY56 A0A0P5ZC73 A0A0P5LQ75 A0A0P6B4X8 A0A0P6CSR3 A0A0P6DFV0
Ontologies
GO
PANTHER
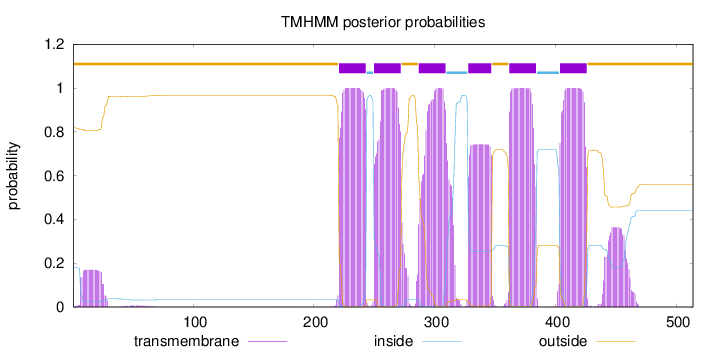
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.809135
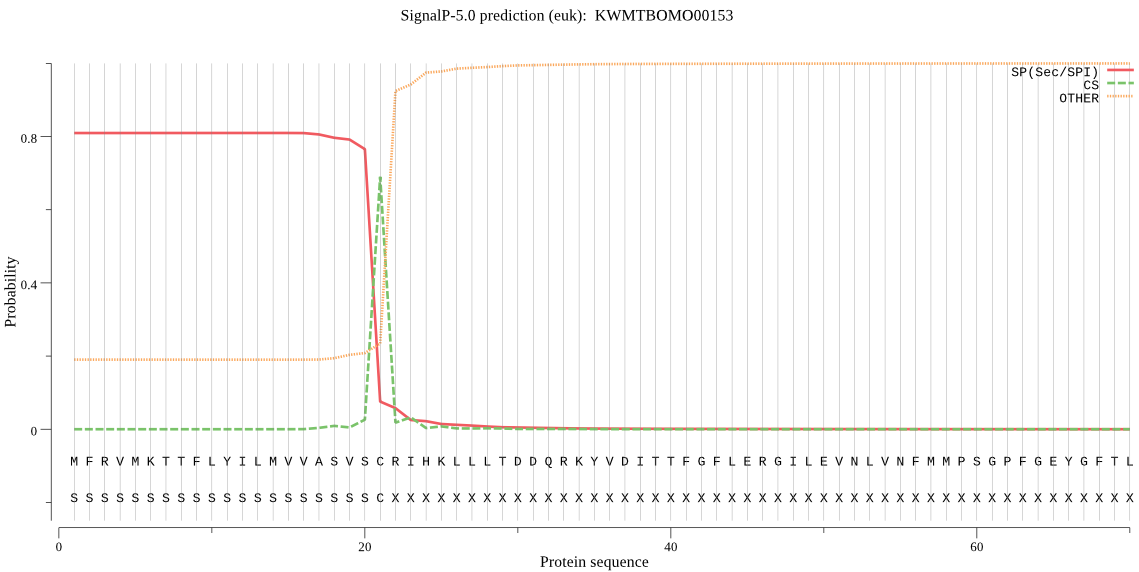
Length:
514
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
138.15391
Exp number, first 60 AAs:
3.4313
Total prob of N-in:
0.18192
outside
1 - 220
TMhelix
221 - 243
inside
244 - 249
TMhelix
250 - 272
outside
273 - 286
TMhelix
287 - 309
inside
310 - 327
TMhelix
328 - 347
outside
348 - 361
TMhelix
362 - 384
inside
385 - 403
TMhelix
404 - 426
outside
427 - 514
Population Genetic Test Statistics
Pi
5.569947
Theta
17.903924
Tajima's D
-2.095382
CLR
0.003059
CSRT
0.00834958252087396
Interpretation
Uncertain
