Gene
KWMTBOMO00147 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002004
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_F_member_2_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.021 Nuclear Reliability : 1.888
Sequence
CDS
ATGCCGTCGGACGCGAAGAAAAGGGCTCAGCAGAAGAAGAAGGAGCAAGCTAGCGGCAGAAACAAGAAGCCGGTGGTTCGTGTGACGAACGAAGACGAAAATGGTATCGGAACTAATGGTATATCCAGGGAAGACCGTGAGCTGACGGCAGAAGAGGCATTGTGCCTTAAGCTAGAAGCTGAAGCGAAAATGAATTCTGAAGCTCGTTCTTGTACTGGGTCACTGGCTGTGCACCCTCGTTCTAGAGATATCAAGATAGCTAACTTTTCAATTACTTTCTATGGGAGTGAGCTACTTCAAGATACTCTCCTGGAGTTGAATTGTGGGCGAAGATATGGGCTAGTCGGACTTAATGGGTGTGGAAAATCATCATTGCTTGCTGCCTTAGGACGCCGTGAGGTTCCTATTCCGGAACACATTGACATATTCCATCTGACTCGAGAAATGCCTGCTTCAGATAAGACTGCACTTCAGTGCGTCATGGAGGTTGACGAGGAGAGAGTCAAACTCGAGAAGCTCGCAGAAATGCTAGCCCACTGTGAAGATGATGAATCACAGGAGCAATTGATGGATGTATACGATCGTCTGGACGATTTGAGCGCCGACACGGCAGAGGCGCGTGCTGCTAATATCCTGCATGGTCTTGGCTTCACCAAGGAAATGCAGCAGAAGGCAACGAAAGACTTCTCCGGAGGTTGGCGTATGCGTATAGCTCTGGCACGAGCGCTATATGTTAAGCCGCACTTACTGCTGCTGGACGAGCCAACAAACCATCTCGATTTGGACGCCTGTGTGTGGCTTGAAGAAGAACTTAAACACTACAAAAGAATTTTAGTGTTGATCTCGCATTCGCAAGACTTCTTGAACGGTGTATGCACGAATATCGTTCATATGAGTAAGCGCCGTTTGAAGTATTACACTGGTAACTATGAAGCCTTCGTTAGGACGCGCATGGAATTGCTAGAGAATCAGATGAAACAGTACAACTGGGAACAAGACCAGATTGCACATATGAAAAACTACATAGCGAGATTCGGTCATGGTTCCGCCAAATTGGCGCGTCAAGCACAGTCCAAAGAAAAGACGTTAGCCAAAATGGTGGCACAGGGTCTCACCGAGAAGGTGGTTGATGACAAGATACTCAATTTCTACTTTCCGTCTTGTGGGAAAGTACCGCCACCCGTCATCATGGTGCAGAATGTATCATTCAGATACACTGATTCTGGGCCGTGGATATACAAGAATTTGGAATTTGGCATTGACCTTGATACAAGACTTGCTTTGGTTGGACCCAATGGAGCAGGCAAGTCAACGCTACTCAAGCTGCTTTATGGAGATTTAGTGCCTTCGACTGGTATGATCCGTAAAAACTCACATCTTCGTATTGGTCGTTACCACCAACACTTGCACGAGTTGTTGGATCTCGATCTGTCTCCACTTGATTACATGATGAAAGAGTTTCCCGAGGTTCGTGAGCGCGAAGAAATGAGAAAAATCATCGGTCGGTATGGTCTCACTGGGAGGCAGCAGGTGTGTCCCATGCGTCAGTTATCCGACGGGCAGCGTTGTCGTGTTGTATTCGCGTGGTTGGCCTGGCAGACTCCGCACCTGCTGTTATTGGACGAGCCCACTAACCATCTTGATATGGAAACCATCGATGCACTCGCAGACGCCATTAACGACTTTGACGGTGGCATGGTGCTCGTTAGTCACGACTTTAGACTTATTAATCAGGTGGCGAATGAAATTTGGATCTGTGAGAACGGTACTACCACAAAATGGGAGGGTGGTATCCTCAAATACAAAGATCACCTGAAATCTAAGATATTGAAGGATAATGCGGATAGCGCCGCTAAAATACGCAAATAG
Protein
MPSDAKKRAQQKKKEQASGRNKKPVVRVTNEDENGIGTNGISREDRELTAEEALCLKLEAEAKMNSEARSCTGSLAVHPRSRDIKIANFSITFYGSELLQDTLLELNCGRRYGLVGLNGCGKSSLLAALGRREVPIPEHIDIFHLTREMPASDKTALQCVMEVDEERVKLEKLAEMLAHCEDDESQEQLMDVYDRLDDLSADTAEARAANILHGLGFTKEMQQKATKDFSGGWRMRIALARALYVKPHLLLLDEPTNHLDLDACVWLEEELKHYKRILVLISHSQDFLNGVCTNIVHMSKRRLKYYTGNYEAFVRTRMELLENQMKQYNWEQDQIAHMKNYIARFGHGSAKLARQAQSKEKTLAKMVAQGLTEKVVDDKILNFYFPSCGKVPPPVIMVQNVSFRYTDSGPWIYKNLEFGIDLDTRLALVGPNGAGKSTLLKLLYGDLVPSTGMIRKNSHLRIGRYHQHLHELLDLDLSPLDYMMKEFPEVREREEMRKIIGRYGLTGRQQVCPMRQLSDGQRCRVVFAWLAWQTPHLLLLDEPTNHLDMETIDALADAINDFDGGMVLVSHDFRLINQVANEIWICENGTTTKWEGGILKYKDHLKSKILKDNADSAAKIRK
Summary
Uniprot
H9IXM1
Q1HQD2
A0A2H4KAX7
A0A2H1VII9
A0A212FHJ1
A0A194QVX9
+ More
A0A194Q6F6 A0A2A4JDW4 A0A2Z5U7C0 A0A0A1X3I6 D6WNA4 W8C527 A0A034VMA5 A0A1L8DFA7 A0A0K8TPF1 A0A1L8DFG7 A0A0K8V3D6 B3MZ10 A0A0U9HSF6 B4L701 B4MA76 B4JX52 B4R5G2 A0A3L8DF04 A0A1W4V3U7 A0A1B6CXJ3 B4PWR4 B4MJ49 B4IKA8 A0A026VTK1 Q9VXR5 A0A1B6DLF1 A0A1S3CVD4 B3NTH6 A0A1B0FDX9 A0A3B0JEH6 A0A1B0AN90 A0A1A9XH56 A0A1A9VRS1 Q29IA7 K7IM55 A0A1A9ZIC2 A0A0R3NZB2 B4GWB0 A0A1B6FHU5 A0A232FCH9 A0A1A9WSY0 A0A161UF68 A0A1L8ECL4 A0A1B6JD99 A0A1I8NWP8 A0A0C9PUT3 J3JYW4 T1P9Q4 A0A2J7QKQ3 A0A0M9A0L2 A0A1B6KIP0 A0A084WR87 A0A067R4B8 A0A0M3QZK8 T1E778 A0A182JHZ5 A0A2M3ZZ78 B0W6T7 A0A1Q3FEN3 A0A2A3ENJ5 A0A087ZP04 A0A1J1IW90 A0A310SAH6 A0A182XAF6 Q7PQX1 A0A182HS95 A0A182VHJ8 A0A182LTQ8 A0A182NR73 A0A182VZV8 A0A182RTI0 Q16RE9 A0A0L0C710 W5JST3 E2BI03 A0A023EUB4 A0A182FQ41 A0A182PKT3 A0A182JNN8 U5ETJ0 A0A182Y419 A0A182QSR7 E0VXE8 E2AF36 A0A158NAA0 E9I9F3 A0A0L7R0I8 F4WJU9 A0A0A9Z993 A0A195BCG3 T1HZ13 A0A195FTP6 J9K7W5 A0A0V0G2N8
A0A194Q6F6 A0A2A4JDW4 A0A2Z5U7C0 A0A0A1X3I6 D6WNA4 W8C527 A0A034VMA5 A0A1L8DFA7 A0A0K8TPF1 A0A1L8DFG7 A0A0K8V3D6 B3MZ10 A0A0U9HSF6 B4L701 B4MA76 B4JX52 B4R5G2 A0A3L8DF04 A0A1W4V3U7 A0A1B6CXJ3 B4PWR4 B4MJ49 B4IKA8 A0A026VTK1 Q9VXR5 A0A1B6DLF1 A0A1S3CVD4 B3NTH6 A0A1B0FDX9 A0A3B0JEH6 A0A1B0AN90 A0A1A9XH56 A0A1A9VRS1 Q29IA7 K7IM55 A0A1A9ZIC2 A0A0R3NZB2 B4GWB0 A0A1B6FHU5 A0A232FCH9 A0A1A9WSY0 A0A161UF68 A0A1L8ECL4 A0A1B6JD99 A0A1I8NWP8 A0A0C9PUT3 J3JYW4 T1P9Q4 A0A2J7QKQ3 A0A0M9A0L2 A0A1B6KIP0 A0A084WR87 A0A067R4B8 A0A0M3QZK8 T1E778 A0A182JHZ5 A0A2M3ZZ78 B0W6T7 A0A1Q3FEN3 A0A2A3ENJ5 A0A087ZP04 A0A1J1IW90 A0A310SAH6 A0A182XAF6 Q7PQX1 A0A182HS95 A0A182VHJ8 A0A182LTQ8 A0A182NR73 A0A182VZV8 A0A182RTI0 Q16RE9 A0A0L0C710 W5JST3 E2BI03 A0A023EUB4 A0A182FQ41 A0A182PKT3 A0A182JNN8 U5ETJ0 A0A182Y419 A0A182QSR7 E0VXE8 E2AF36 A0A158NAA0 E9I9F3 A0A0L7R0I8 F4WJU9 A0A0A9Z993 A0A195BCG3 T1HZ13 A0A195FTP6 J9K7W5 A0A0V0G2N8
Pubmed
19121390
22118469
26354079
25830018
18362917
19820115
+ More
24495485 25348373 29121518 26369729 17994087 18057021 30249741 17550304 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20075255 23185243 28648823 22516182 23537049 25315136 24438588 24845553 12364791 14747013 17210077 17510324 26108605 20920257 23761445 20798317 24945155 26483478 25244985 20566863 21347285 21282665 21719571 25401762 26823975
24495485 25348373 29121518 26369729 17994087 18057021 30249741 17550304 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20075255 23185243 28648823 22516182 23537049 25315136 24438588 24845553 12364791 14747013 17210077 17510324 26108605 20920257 23761445 20798317 24945155 26483478 25244985 20566863 21347285 21282665 21719571 25401762 26823975
EMBL
BABH01016003
DQ443120
ABF51209.1
KY484785
ASS36027.1
ODYU01002555
+ More
SOQ40212.1 AGBW02008503 OWR53209.1 KQ461108 KPJ09120.1 KQ459463 KPJ00585.1 NWSH01001858 PCG69898.1 AP017515 BBB06849.1 GBXI01009059 JAD05233.1 KQ971342 EFA03773.1 GAMC01005124 JAC01432.1 KY849636 GAKP01015348 ATY74517.1 JAC43604.1 GFDF01008966 JAV05118.1 GDAI01001336 JAI16267.1 GFDF01008967 JAV05117.1 GDHF01019209 GDHF01016348 JAI33105.1 JAI35966.1 CH902632 EDV32854.1 KPU74167.1 GARF01000066 JAC88892.1 CH933812 EDW06147.1 KRG07163.1 CH940655 EDW66135.1 CH916376 EDV95328.1 CM000366 EDX18007.1 QOIP01000009 RLU18469.1 GEDC01019119 GEDC01004094 JAS18179.1 JAS33204.1 CM000162 EDX01810.1 CH963738 EDW72138.1 CH480852 EDW51500.1 KK108171 EZA46836.1 AE014298 AY061059 AAF48493.1 AAL28607.1 GEDC01027856 GEDC01026318 GEDC01016806 GEDC01010803 GEDC01009050 GEDC01002643 JAS09442.1 JAS10980.1 JAS20492.1 JAS26495.1 JAS28248.1 JAS34655.1 MH172508 QAA95915.1 CH954180 EDV46980.1 CCAG010011618 OUUW01000003 SPP78582.1 JXJN01000753 CH379063 EAL32746.2 KRT06281.1 KRT06282.1 CH479194 EDW26994.1 GECZ01019993 JAS49776.1 NNAY01000419 OXU28531.1 KF828792 AIN44109.1 GFDG01002352 JAV16447.1 GECU01010608 JAS97098.1 GBYB01005168 JAG74935.1 APGK01037599 BT128445 KB740948 KB632288 AEE63402.1 ENN77364.1 ERL91490.1 KA645476 AFP60105.1 NEVH01013262 PNF29138.1 KQ435798 KOX73379.1 GEBQ01028655 JAT11322.1 ATLV01025994 KE525404 KFB52731.1 KK852902 KDR14063.1 CP012528 ALC49593.1 GAMD01003282 JAA98308.1 GGFK01000461 MBW33782.1 DS231849 EDS36918.1 GFDL01009037 JAV26008.1 KZ288204 PBC33270.1 CVRI01000063 CRL04477.1 KQ762329 OAD55974.1 AAAB01008859 EAA08160.6 APCN01000256 AXCM01000275 CH477714 EAT36980.1 JRES01000814 KNC28193.1 ADMH02000547 ETN65975.1 GL448407 EFN84681.1 JXUM01100955 GAPW01000598 KQ564612 JAC13000.1 KXJ72019.1 GANO01002757 JAB57114.1 AXCN02001469 DS235830 EEB18054.1 GL438984 EFN67991.1 ADTU01001463 ADTU01001464 GL761800 EFZ22792.1 KQ414670 KOC64362.1 GL888186 EGI65591.1 GBHO01005224 GBRD01010341 GBRD01010340 GDHC01013851 GDHC01009732 JAG38380.1 JAG55483.1 JAQ04778.1 JAQ08897.1 KQ976528 KYM81885.1 ACPB03021331 KQ981264 KYN43990.1 ABLF02027288 GECL01003743 JAP02381.1
SOQ40212.1 AGBW02008503 OWR53209.1 KQ461108 KPJ09120.1 KQ459463 KPJ00585.1 NWSH01001858 PCG69898.1 AP017515 BBB06849.1 GBXI01009059 JAD05233.1 KQ971342 EFA03773.1 GAMC01005124 JAC01432.1 KY849636 GAKP01015348 ATY74517.1 JAC43604.1 GFDF01008966 JAV05118.1 GDAI01001336 JAI16267.1 GFDF01008967 JAV05117.1 GDHF01019209 GDHF01016348 JAI33105.1 JAI35966.1 CH902632 EDV32854.1 KPU74167.1 GARF01000066 JAC88892.1 CH933812 EDW06147.1 KRG07163.1 CH940655 EDW66135.1 CH916376 EDV95328.1 CM000366 EDX18007.1 QOIP01000009 RLU18469.1 GEDC01019119 GEDC01004094 JAS18179.1 JAS33204.1 CM000162 EDX01810.1 CH963738 EDW72138.1 CH480852 EDW51500.1 KK108171 EZA46836.1 AE014298 AY061059 AAF48493.1 AAL28607.1 GEDC01027856 GEDC01026318 GEDC01016806 GEDC01010803 GEDC01009050 GEDC01002643 JAS09442.1 JAS10980.1 JAS20492.1 JAS26495.1 JAS28248.1 JAS34655.1 MH172508 QAA95915.1 CH954180 EDV46980.1 CCAG010011618 OUUW01000003 SPP78582.1 JXJN01000753 CH379063 EAL32746.2 KRT06281.1 KRT06282.1 CH479194 EDW26994.1 GECZ01019993 JAS49776.1 NNAY01000419 OXU28531.1 KF828792 AIN44109.1 GFDG01002352 JAV16447.1 GECU01010608 JAS97098.1 GBYB01005168 JAG74935.1 APGK01037599 BT128445 KB740948 KB632288 AEE63402.1 ENN77364.1 ERL91490.1 KA645476 AFP60105.1 NEVH01013262 PNF29138.1 KQ435798 KOX73379.1 GEBQ01028655 JAT11322.1 ATLV01025994 KE525404 KFB52731.1 KK852902 KDR14063.1 CP012528 ALC49593.1 GAMD01003282 JAA98308.1 GGFK01000461 MBW33782.1 DS231849 EDS36918.1 GFDL01009037 JAV26008.1 KZ288204 PBC33270.1 CVRI01000063 CRL04477.1 KQ762329 OAD55974.1 AAAB01008859 EAA08160.6 APCN01000256 AXCM01000275 CH477714 EAT36980.1 JRES01000814 KNC28193.1 ADMH02000547 ETN65975.1 GL448407 EFN84681.1 JXUM01100955 GAPW01000598 KQ564612 JAC13000.1 KXJ72019.1 GANO01002757 JAB57114.1 AXCN02001469 DS235830 EEB18054.1 GL438984 EFN67991.1 ADTU01001463 ADTU01001464 GL761800 EFZ22792.1 KQ414670 KOC64362.1 GL888186 EGI65591.1 GBHO01005224 GBRD01010341 GBRD01010340 GDHC01013851 GDHC01009732 JAG38380.1 JAG55483.1 JAQ04778.1 JAQ08897.1 KQ976528 KYM81885.1 ACPB03021331 KQ981264 KYN43990.1 ABLF02027288 GECL01003743 JAP02381.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000218220
UP000007266
+ More
UP000007801 UP000009192 UP000008792 UP000001070 UP000000304 UP000279307 UP000192221 UP000002282 UP000007798 UP000001292 UP000053097 UP000000803 UP000079169 UP000008711 UP000092444 UP000268350 UP000092460 UP000092443 UP000078200 UP000001819 UP000002358 UP000092445 UP000008744 UP000215335 UP000091820 UP000095300 UP000019118 UP000030742 UP000095301 UP000235965 UP000053105 UP000030765 UP000027135 UP000092553 UP000075880 UP000002320 UP000242457 UP000005203 UP000183832 UP000076407 UP000007062 UP000075840 UP000075903 UP000075883 UP000075884 UP000075920 UP000075900 UP000008820 UP000037069 UP000000673 UP000008237 UP000069940 UP000249989 UP000069272 UP000075885 UP000075881 UP000076408 UP000075886 UP000009046 UP000000311 UP000005205 UP000053825 UP000007755 UP000078540 UP000015103 UP000078541 UP000007819
UP000007801 UP000009192 UP000008792 UP000001070 UP000000304 UP000279307 UP000192221 UP000002282 UP000007798 UP000001292 UP000053097 UP000000803 UP000079169 UP000008711 UP000092444 UP000268350 UP000092460 UP000092443 UP000078200 UP000001819 UP000002358 UP000092445 UP000008744 UP000215335 UP000091820 UP000095300 UP000019118 UP000030742 UP000095301 UP000235965 UP000053105 UP000030765 UP000027135 UP000092553 UP000075880 UP000002320 UP000242457 UP000005203 UP000183832 UP000076407 UP000007062 UP000075840 UP000075903 UP000075883 UP000075884 UP000075920 UP000075900 UP000008820 UP000037069 UP000000673 UP000008237 UP000069940 UP000249989 UP000069272 UP000075885 UP000075881 UP000076408 UP000075886 UP000009046 UP000000311 UP000005205 UP000053825 UP000007755 UP000078540 UP000015103 UP000078541 UP000007819
Interpro
Gene 3D
ProteinModelPortal
H9IXM1
Q1HQD2
A0A2H4KAX7
A0A2H1VII9
A0A212FHJ1
A0A194QVX9
+ More
A0A194Q6F6 A0A2A4JDW4 A0A2Z5U7C0 A0A0A1X3I6 D6WNA4 W8C527 A0A034VMA5 A0A1L8DFA7 A0A0K8TPF1 A0A1L8DFG7 A0A0K8V3D6 B3MZ10 A0A0U9HSF6 B4L701 B4MA76 B4JX52 B4R5G2 A0A3L8DF04 A0A1W4V3U7 A0A1B6CXJ3 B4PWR4 B4MJ49 B4IKA8 A0A026VTK1 Q9VXR5 A0A1B6DLF1 A0A1S3CVD4 B3NTH6 A0A1B0FDX9 A0A3B0JEH6 A0A1B0AN90 A0A1A9XH56 A0A1A9VRS1 Q29IA7 K7IM55 A0A1A9ZIC2 A0A0R3NZB2 B4GWB0 A0A1B6FHU5 A0A232FCH9 A0A1A9WSY0 A0A161UF68 A0A1L8ECL4 A0A1B6JD99 A0A1I8NWP8 A0A0C9PUT3 J3JYW4 T1P9Q4 A0A2J7QKQ3 A0A0M9A0L2 A0A1B6KIP0 A0A084WR87 A0A067R4B8 A0A0M3QZK8 T1E778 A0A182JHZ5 A0A2M3ZZ78 B0W6T7 A0A1Q3FEN3 A0A2A3ENJ5 A0A087ZP04 A0A1J1IW90 A0A310SAH6 A0A182XAF6 Q7PQX1 A0A182HS95 A0A182VHJ8 A0A182LTQ8 A0A182NR73 A0A182VZV8 A0A182RTI0 Q16RE9 A0A0L0C710 W5JST3 E2BI03 A0A023EUB4 A0A182FQ41 A0A182PKT3 A0A182JNN8 U5ETJ0 A0A182Y419 A0A182QSR7 E0VXE8 E2AF36 A0A158NAA0 E9I9F3 A0A0L7R0I8 F4WJU9 A0A0A9Z993 A0A195BCG3 T1HZ13 A0A195FTP6 J9K7W5 A0A0V0G2N8
A0A194Q6F6 A0A2A4JDW4 A0A2Z5U7C0 A0A0A1X3I6 D6WNA4 W8C527 A0A034VMA5 A0A1L8DFA7 A0A0K8TPF1 A0A1L8DFG7 A0A0K8V3D6 B3MZ10 A0A0U9HSF6 B4L701 B4MA76 B4JX52 B4R5G2 A0A3L8DF04 A0A1W4V3U7 A0A1B6CXJ3 B4PWR4 B4MJ49 B4IKA8 A0A026VTK1 Q9VXR5 A0A1B6DLF1 A0A1S3CVD4 B3NTH6 A0A1B0FDX9 A0A3B0JEH6 A0A1B0AN90 A0A1A9XH56 A0A1A9VRS1 Q29IA7 K7IM55 A0A1A9ZIC2 A0A0R3NZB2 B4GWB0 A0A1B6FHU5 A0A232FCH9 A0A1A9WSY0 A0A161UF68 A0A1L8ECL4 A0A1B6JD99 A0A1I8NWP8 A0A0C9PUT3 J3JYW4 T1P9Q4 A0A2J7QKQ3 A0A0M9A0L2 A0A1B6KIP0 A0A084WR87 A0A067R4B8 A0A0M3QZK8 T1E778 A0A182JHZ5 A0A2M3ZZ78 B0W6T7 A0A1Q3FEN3 A0A2A3ENJ5 A0A087ZP04 A0A1J1IW90 A0A310SAH6 A0A182XAF6 Q7PQX1 A0A182HS95 A0A182VHJ8 A0A182LTQ8 A0A182NR73 A0A182VZV8 A0A182RTI0 Q16RE9 A0A0L0C710 W5JST3 E2BI03 A0A023EUB4 A0A182FQ41 A0A182PKT3 A0A182JNN8 U5ETJ0 A0A182Y419 A0A182QSR7 E0VXE8 E2AF36 A0A158NAA0 E9I9F3 A0A0L7R0I8 F4WJU9 A0A0A9Z993 A0A195BCG3 T1HZ13 A0A195FTP6 J9K7W5 A0A0V0G2N8
PDB
5ZXD
E-value=3.45928e-95,
Score=891
Ontologies
GO
Topology
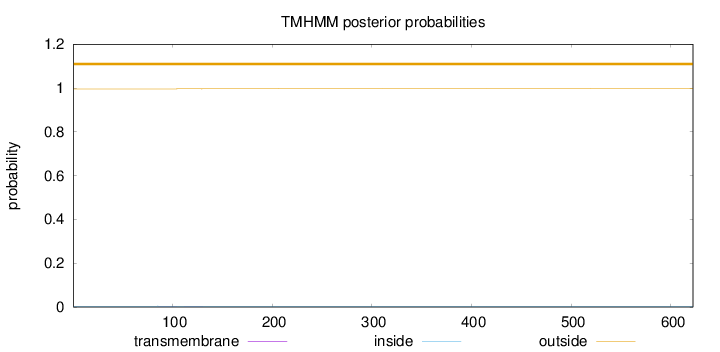
Length:
622
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0176399999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00399
outside
1 - 622
Population Genetic Test Statistics
Pi
11.558453
Theta
12.32763
Tajima's D
-0.658651
CLR
0.005579
CSRT
0.206889655517224
Interpretation
Possibly Positive selection
