Gene
KWMTBOMO00146 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002005
Annotation
PREDICTED:_ADP?ATP_carrier_protein-like_[Amyelois_transitella]
Full name
ADP,ATP carrier protein
Alternative Name
ADP/ATP translocase
Adenine nucleotide translocator
Stress-sensitive protein B
Adenine nucleotide translocator
Stress-sensitive protein B
Location in the cell
Mitochondrial Reliability : 1.224 PlasmaMembrane Reliability : 1.368
Sequence
CDS
ATGTCGGATAAAGATAAAAAGAAGGGTCCAAAGAAAGAGGATGAAGCAAGCTATGGTTTTTTAAAAGATTTTTTAGCCGGTGGAATTTCTGCTGCTATATCGAAAACAGCAGTTGCTCCAATTGAAAGAGTTAAACTTATTTTACAAGTTCAACATGTGTCCAAACAGATATCAGAAGACAAAAGATATAAAGGCATGGTGGACGCATTTGTCCGAATACCTAAAGAACAAGGTTTTTTGTCTTTCTGGCGAGGAAACTTGGCTAACGTAATACGGTACTTTCCGACACAAGCGCTGAATTTTGCTTTCAAAGACGTGTACAAAGGCATATTTTTAGAGGGAGTAGATAAGAACAAGCAGTTTTGGCGTCATTTTGCTGGAAATTTAGCTTCAGGAGGTGCTGCTGGAGCAACATCGCTATGCTTCGTATATCCGTTGGATTTTGCTAGAACAAGATTAGCAGCTGACGTGGGAAAGGGCAAGGACAAGGAATTTACAGGGCTGGTGAACTGTCTGATCAAGACACTCAAGTCCGACGGTCCTATGGGGCTTTATCGAGGCTTCGTAGTGTCCGTTCAGGGCATCATCATCTACCGCGCCACATACTTCGGATGCTTTGATACGGCGCGCGACATGTTACCCGATCCTAAGAATACATCGCTTCTTATTACTTGGATGATTGCTCAGACTGTAACTACAGTGGCCGGTATTGCGTCGTATCCCTTGGACACTGTCCGTCGTCGAATGATGATGCAGTCCGGACGACCACTCAATGAGCGCCAGTACAAGAGCACGGCGCATTGTTGGGCGACCATCCTCAAAACGGAGGGAGCCGGTGCTTTCTTTAAGGGCGCCTTCTCGAATGTCCTGAGAGGCACGGGAGGAGCGTTTGTGCTTGTCTTGTATGATGAAATAAAAAAAGTATTATAG
Protein
MSDKDKKKGPKKEDEASYGFLKDFLAGGISAAISKTAVAPIERVKLILQVQHVSKQISEDKRYKGMVDAFVRIPKEQGFLSFWRGNLANVIRYFPTQALNFAFKDVYKGIFLEGVDKNKQFWRHFAGNLASGGAAGATSLCFVYPLDFARTRLAADVGKGKDKEFTGLVNCLIKTLKSDGPMGLYRGFVVSVQGIIIYRATYFGCFDTARDMLPDPKNTSLLITWMIAQTVTTVAGIASYPLDTVRRRMMMQSGRPLNERQYKSTAHCWATILKTEGAGAFFKGAFSNVLRGTGGAFVLVLYDEIKKVL
Summary
Subunit
Homodimer.
Similarity
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Keywords
Alternative splicing
Complete proteome
Membrane
Mitochondrion
Mitochondrion inner membrane
Reference proteome
Repeat
Transmembrane
Transmembrane helix
Transport
Feature
chain ADP,ATP carrier protein
splice variant In isoform A.
splice variant In isoform A.
Uniprot
H9IXM2
A0A212FHI8
A0A194QUX6
A0A0E4FHB5
A0A194Q5F3
A0A0E4AY98
+ More
A0A0A1X6J4 A0A0X7Z554 A0A034WNL1 A0A0C9Q032 A0A212FB24 W8BQQ1 D1Z385 Q26365 B4H0G1 Q29H33 Q26365-2 B4R7M9 X2JB48 B4IDU1 A0A2A4JPR6 V5H0R0 Q86PT7 A0A1W4V0V8 D0QWE8 B3NVE5 B4PXZ8 A0A3B0K5A9 A0A023GKH9 I4DII1 I4DMD7 B7Q4Q4 A7KCY1 A0A3G1T198 C7B401 A0A0M4F069 A0A2R5L5E1 A0A1L8EFF9 Q9NHW5 A0A1I8PSD6 S4P9Q1 B4L7X9 T1PEW0 B4JK92 A0A023FHD0 A0A1A9WA72 A0A1B0FQ42 A0A0L7KEA5 Q86CZ0 A0A1A9XKG5 A0A1A9VNE9 A0A1B0AUC4 A0A1B0AFE0 D3TNQ0 A0A194R4M6 A0A1L8EAW7 A0A0K8TQ79 Q86PG2 A0A1L8EBM1 A0A2A4J0S3 B4M9X6 B4N1K0 T1KJF8 A0A0E3VJG9 B3MQD5 A0A2H1WC32 A0A0C9Q2V5 A0A232EPC8 K7IW87 A0A0T6BBP8 A7XQS5 A0A224YZR0 Q4PMB2 N6TIB1 V5JDG5 E0W200 A0A0L7QZ33 Q6VQ13 A0A0E4AY35 B4IDU0 A0A0K8R6D7 B4N1J9 D6WIT1 A0A0E4AXA9 A0A131YNP5 B3NVE6 O62526 A2ICN6 A0A2A3ELD5 V9IK47 A0A023GL78 A0A2I9LPW0 A0A2J7NKC2 A0A1W4UPD0 A0A0C9R2A0 A0A147BVU8 A0A2S2PZ59 Q5XXT2 A0A1W7R9D7 T1KJF7
A0A0A1X6J4 A0A0X7Z554 A0A034WNL1 A0A0C9Q032 A0A212FB24 W8BQQ1 D1Z385 Q26365 B4H0G1 Q29H33 Q26365-2 B4R7M9 X2JB48 B4IDU1 A0A2A4JPR6 V5H0R0 Q86PT7 A0A1W4V0V8 D0QWE8 B3NVE5 B4PXZ8 A0A3B0K5A9 A0A023GKH9 I4DII1 I4DMD7 B7Q4Q4 A7KCY1 A0A3G1T198 C7B401 A0A0M4F069 A0A2R5L5E1 A0A1L8EFF9 Q9NHW5 A0A1I8PSD6 S4P9Q1 B4L7X9 T1PEW0 B4JK92 A0A023FHD0 A0A1A9WA72 A0A1B0FQ42 A0A0L7KEA5 Q86CZ0 A0A1A9XKG5 A0A1A9VNE9 A0A1B0AUC4 A0A1B0AFE0 D3TNQ0 A0A194R4M6 A0A1L8EAW7 A0A0K8TQ79 Q86PG2 A0A1L8EBM1 A0A2A4J0S3 B4M9X6 B4N1K0 T1KJF8 A0A0E3VJG9 B3MQD5 A0A2H1WC32 A0A0C9Q2V5 A0A232EPC8 K7IW87 A0A0T6BBP8 A7XQS5 A0A224YZR0 Q4PMB2 N6TIB1 V5JDG5 E0W200 A0A0L7QZ33 Q6VQ13 A0A0E4AY35 B4IDU0 A0A0K8R6D7 B4N1J9 D6WIT1 A0A0E4AXA9 A0A131YNP5 B3NVE6 O62526 A2ICN6 A0A2A3ELD5 V9IK47 A0A023GL78 A0A2I9LPW0 A0A2J7NKC2 A0A1W4UPD0 A0A0C9R2A0 A0A147BVU8 A0A2S2PZ59 Q5XXT2 A0A1W7R9D7 T1KJF7
Pubmed
19121390
25742135
22118469
26354079
25830018
25348373
+ More
24495485 1387687 7520869 10511565 10731132 12537572 12537569 17994087 15632085 23185243 12537568 12537573 12537574 16110336 17569856 17569867 25765539 19859648 18057021 17550304 22651552 17603110 23622113 25315136 26227816 28756777 20353571 26369729 28648823 20075255 28797301 16431279 23537049 20566863 18362917 19820115 26830274 26109357 26109356 29248469 26131772
24495485 1387687 7520869 10511565 10731132 12537572 12537569 17994087 15632085 23185243 12537568 12537573 12537574 16110336 17569856 17569867 25765539 19859648 18057021 17550304 22651552 17603110 23622113 25315136 26227816 28756777 20353571 26369729 28648823 20075255 28797301 16431279 23537049 20566863 18362917 19820115 26830274 26109357 26109356 29248469 26131772
EMBL
BABH01016002
AB928003
BAR13230.1
AGBW02008503
OWR53210.1
KQ461108
+ More
KPJ09119.1 AB928008 BAR13235.1 KQ459463 KPJ00584.1 AB928005 BAR13232.1 GBXI01007545 JAD06747.1 KP198608 AKQ44385.1 GAKP01003197 JAC55755.1 GBYB01007193 JAG76960.1 AGBW02009381 OWR50945.1 GAMC01007413 GAMC01007412 JAB99143.1 BT120021 ACZ98473.1 S43651 S71762 Y10618 AE014298 AY060978 AY070894 BT011069 CH479200 EDW29756.1 CH379064 EAL31926.1 KRT06687.1 CM000366 EDX17588.1 AHN59560.1 CH480830 EDW45749.1 NWSH01000864 PCG73809.1 GANP01007893 JAB76575.1 AY186577 AAO32325.1 FJ821026 ACN94645.1 CH954180 EDV46133.1 KQS29842.1 CM000162 EDX02970.1 KRK07048.1 OUUW01000021 SPP89417.1 GBBM01001875 JAC33543.1 AK401099 KQ459580 BAM17721.1 KPI99189.1 AK402455 BAM19077.1 ABJB010565110 ABJB010582667 ABJB010602457 ABJB010639503 ABJB010825073 ABJB011063103 DS857340 EEC13826.1 EF207974 ABS57449.1 MG846914 AXY94766.1 FJ788509 ACT91090.1 CP012528 ALC49980.1 GGLE01000544 MBY04670.1 GFDG01001454 JAV17345.1 AF218587 AAF32322.1 GAIX01003659 JAA88901.1 CH933814 EDW05554.1 KRG07447.1 KA647327 AFP61956.1 CH916370 EDV99994.1 GBBK01003096 JAC21386.1 CCAG010006700 JTDY01010031 KOB61648.1 AY253868 KZ150209 AAP20934.2 PZC72215.1 JXJN01003606 EZ423052 EZ423060 ADD19328.1 ADD19336.1 KQ460685 KPJ12768.1 GFDG01002921 JAV15878.1 GDAI01001307 JAI16296.1 AY227000 AB928002 AAO32817.1 BAR13229.1 GFDG01002668 JAV16131.1 NWSH01004536 PCG64973.1 CH940655 EDW66035.1 KRF82544.1 CH963925 EDW78239.1 KRF98835.1 CAEY01000134 CAEY01000135 CAEY01001799 AB928017 BAR13244.1 AB928007 BAR13234.1 CH902621 EDV44561.1 KPU81637.1 ODYU01007535 SOQ50392.1 GBYB01008368 GBYB01008370 JAG78135.1 JAG78137.1 NNAY01002956 OXU20215.1 LJIG01002415 KRT84525.1 EU093076 ABU68467.1 GFPF01011950 MAA23096.1 DQ066215 AAY66852.1 APGK01036482 KB740941 KB632070 ENN77548.1 ERL88543.1 KC122907 AB928015 AGI96985.1 BAR13242.1 DS235873 EEB19594.1 KQ414683 KOC63879.1 AY332626 AY568009 AAQ24500.1 AAS73299.1 AB928016 BAR13243.1 EDW45748.1 GADI01007091 JAA66717.1 EDW78238.1 KQ971321 EEZ99521.1 AB928006 BAR13233.1 GEDV01008379 JAP80178.1 EDV46134.1 BT044582 BT044583 BT088432 AAF47956.1 AAO41648.1 ACI31282.1 ACI31283.1 ACR40400.1 AGB95275.1 CAA71629.1 EF194157 ABM69092.1 KZ288223 PBC31969.1 JR049134 AEY60871.1 GBBM01001843 JAC33575.1 GFWZ01000424 MBW20414.1 NEVH01054106 PNE09422.1 GBZX01001595 JAG91145.1 GEGO01000544 JAR94860.1 GGMS01001461 MBY70664.1 AY725780 AAU95193.1 GFAH01000639 JAV47750.1
KPJ09119.1 AB928008 BAR13235.1 KQ459463 KPJ00584.1 AB928005 BAR13232.1 GBXI01007545 JAD06747.1 KP198608 AKQ44385.1 GAKP01003197 JAC55755.1 GBYB01007193 JAG76960.1 AGBW02009381 OWR50945.1 GAMC01007413 GAMC01007412 JAB99143.1 BT120021 ACZ98473.1 S43651 S71762 Y10618 AE014298 AY060978 AY070894 BT011069 CH479200 EDW29756.1 CH379064 EAL31926.1 KRT06687.1 CM000366 EDX17588.1 AHN59560.1 CH480830 EDW45749.1 NWSH01000864 PCG73809.1 GANP01007893 JAB76575.1 AY186577 AAO32325.1 FJ821026 ACN94645.1 CH954180 EDV46133.1 KQS29842.1 CM000162 EDX02970.1 KRK07048.1 OUUW01000021 SPP89417.1 GBBM01001875 JAC33543.1 AK401099 KQ459580 BAM17721.1 KPI99189.1 AK402455 BAM19077.1 ABJB010565110 ABJB010582667 ABJB010602457 ABJB010639503 ABJB010825073 ABJB011063103 DS857340 EEC13826.1 EF207974 ABS57449.1 MG846914 AXY94766.1 FJ788509 ACT91090.1 CP012528 ALC49980.1 GGLE01000544 MBY04670.1 GFDG01001454 JAV17345.1 AF218587 AAF32322.1 GAIX01003659 JAA88901.1 CH933814 EDW05554.1 KRG07447.1 KA647327 AFP61956.1 CH916370 EDV99994.1 GBBK01003096 JAC21386.1 CCAG010006700 JTDY01010031 KOB61648.1 AY253868 KZ150209 AAP20934.2 PZC72215.1 JXJN01003606 EZ423052 EZ423060 ADD19328.1 ADD19336.1 KQ460685 KPJ12768.1 GFDG01002921 JAV15878.1 GDAI01001307 JAI16296.1 AY227000 AB928002 AAO32817.1 BAR13229.1 GFDG01002668 JAV16131.1 NWSH01004536 PCG64973.1 CH940655 EDW66035.1 KRF82544.1 CH963925 EDW78239.1 KRF98835.1 CAEY01000134 CAEY01000135 CAEY01001799 AB928017 BAR13244.1 AB928007 BAR13234.1 CH902621 EDV44561.1 KPU81637.1 ODYU01007535 SOQ50392.1 GBYB01008368 GBYB01008370 JAG78135.1 JAG78137.1 NNAY01002956 OXU20215.1 LJIG01002415 KRT84525.1 EU093076 ABU68467.1 GFPF01011950 MAA23096.1 DQ066215 AAY66852.1 APGK01036482 KB740941 KB632070 ENN77548.1 ERL88543.1 KC122907 AB928015 AGI96985.1 BAR13242.1 DS235873 EEB19594.1 KQ414683 KOC63879.1 AY332626 AY568009 AAQ24500.1 AAS73299.1 AB928016 BAR13243.1 EDW45748.1 GADI01007091 JAA66717.1 EDW78238.1 KQ971321 EEZ99521.1 AB928006 BAR13233.1 GEDV01008379 JAP80178.1 EDV46134.1 BT044582 BT044583 BT088432 AAF47956.1 AAO41648.1 ACI31282.1 ACI31283.1 ACR40400.1 AGB95275.1 CAA71629.1 EF194157 ABM69092.1 KZ288223 PBC31969.1 JR049134 AEY60871.1 GBBM01001843 JAC33575.1 GFWZ01000424 MBW20414.1 NEVH01054106 PNE09422.1 GBZX01001595 JAG91145.1 GEGO01000544 JAR94860.1 GGMS01001461 MBY70664.1 AY725780 AAU95193.1 GFAH01000639 JAV47750.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000000803
UP000008744
+ More
UP000001819 UP000000304 UP000001292 UP000218220 UP000192221 UP000008711 UP000002282 UP000268350 UP000001555 UP000092553 UP000095300 UP000009192 UP000095301 UP000001070 UP000091820 UP000092444 UP000037510 UP000092443 UP000078200 UP000092460 UP000092445 UP000008792 UP000007798 UP000015104 UP000007801 UP000215335 UP000002358 UP000019118 UP000030742 UP000009046 UP000053825 UP000005203 UP000007266 UP000242457 UP000235965
UP000001819 UP000000304 UP000001292 UP000218220 UP000192221 UP000008711 UP000002282 UP000268350 UP000001555 UP000092553 UP000095300 UP000009192 UP000095301 UP000001070 UP000091820 UP000092444 UP000037510 UP000092443 UP000078200 UP000092460 UP000092445 UP000008792 UP000007798 UP000015104 UP000007801 UP000215335 UP000002358 UP000019118 UP000030742 UP000009046 UP000053825 UP000005203 UP000007266 UP000242457 UP000235965
Pfam
PF00153 Mito_carr
Interpro
SUPFAM
SSF103506
SSF103506
Gene 3D
ProteinModelPortal
H9IXM2
A0A212FHI8
A0A194QUX6
A0A0E4FHB5
A0A194Q5F3
A0A0E4AY98
+ More
A0A0A1X6J4 A0A0X7Z554 A0A034WNL1 A0A0C9Q032 A0A212FB24 W8BQQ1 D1Z385 Q26365 B4H0G1 Q29H33 Q26365-2 B4R7M9 X2JB48 B4IDU1 A0A2A4JPR6 V5H0R0 Q86PT7 A0A1W4V0V8 D0QWE8 B3NVE5 B4PXZ8 A0A3B0K5A9 A0A023GKH9 I4DII1 I4DMD7 B7Q4Q4 A7KCY1 A0A3G1T198 C7B401 A0A0M4F069 A0A2R5L5E1 A0A1L8EFF9 Q9NHW5 A0A1I8PSD6 S4P9Q1 B4L7X9 T1PEW0 B4JK92 A0A023FHD0 A0A1A9WA72 A0A1B0FQ42 A0A0L7KEA5 Q86CZ0 A0A1A9XKG5 A0A1A9VNE9 A0A1B0AUC4 A0A1B0AFE0 D3TNQ0 A0A194R4M6 A0A1L8EAW7 A0A0K8TQ79 Q86PG2 A0A1L8EBM1 A0A2A4J0S3 B4M9X6 B4N1K0 T1KJF8 A0A0E3VJG9 B3MQD5 A0A2H1WC32 A0A0C9Q2V5 A0A232EPC8 K7IW87 A0A0T6BBP8 A7XQS5 A0A224YZR0 Q4PMB2 N6TIB1 V5JDG5 E0W200 A0A0L7QZ33 Q6VQ13 A0A0E4AY35 B4IDU0 A0A0K8R6D7 B4N1J9 D6WIT1 A0A0E4AXA9 A0A131YNP5 B3NVE6 O62526 A2ICN6 A0A2A3ELD5 V9IK47 A0A023GL78 A0A2I9LPW0 A0A2J7NKC2 A0A1W4UPD0 A0A0C9R2A0 A0A147BVU8 A0A2S2PZ59 Q5XXT2 A0A1W7R9D7 T1KJF7
A0A0A1X6J4 A0A0X7Z554 A0A034WNL1 A0A0C9Q032 A0A212FB24 W8BQQ1 D1Z385 Q26365 B4H0G1 Q29H33 Q26365-2 B4R7M9 X2JB48 B4IDU1 A0A2A4JPR6 V5H0R0 Q86PT7 A0A1W4V0V8 D0QWE8 B3NVE5 B4PXZ8 A0A3B0K5A9 A0A023GKH9 I4DII1 I4DMD7 B7Q4Q4 A7KCY1 A0A3G1T198 C7B401 A0A0M4F069 A0A2R5L5E1 A0A1L8EFF9 Q9NHW5 A0A1I8PSD6 S4P9Q1 B4L7X9 T1PEW0 B4JK92 A0A023FHD0 A0A1A9WA72 A0A1B0FQ42 A0A0L7KEA5 Q86CZ0 A0A1A9XKG5 A0A1A9VNE9 A0A1B0AUC4 A0A1B0AFE0 D3TNQ0 A0A194R4M6 A0A1L8EAW7 A0A0K8TQ79 Q86PG2 A0A1L8EBM1 A0A2A4J0S3 B4M9X6 B4N1K0 T1KJF8 A0A0E3VJG9 B3MQD5 A0A2H1WC32 A0A0C9Q2V5 A0A232EPC8 K7IW87 A0A0T6BBP8 A7XQS5 A0A224YZR0 Q4PMB2 N6TIB1 V5JDG5 E0W200 A0A0L7QZ33 Q6VQ13 A0A0E4AY35 B4IDU0 A0A0K8R6D7 B4N1J9 D6WIT1 A0A0E4AXA9 A0A131YNP5 B3NVE6 O62526 A2ICN6 A0A2A3ELD5 V9IK47 A0A023GL78 A0A2I9LPW0 A0A2J7NKC2 A0A1W4UPD0 A0A0C9R2A0 A0A147BVU8 A0A2S2PZ59 Q5XXT2 A0A1W7R9D7 T1KJF7
PDB
2C3E
E-value=8.45458e-121,
Score=1108
Ontologies
GO
GO:0022857
GO:0005743
GO:0016021
GO:0051560
GO:0006839
GO:0051480
GO:0051124
GO:0008340
GO:0048489
GO:0045202
GO:0051900
GO:0015866
GO:0040011
GO:0010507
GO:0070050
GO:0005739
GO:0007268
GO:0034599
GO:0001508
GO:0015867
GO:0048477
GO:0005471
GO:0046716
GO:2001171
GO:0005215
GO:0006810
GO:0007018
GO:0030286
GO:0048013
GO:0005003
GO:0005887
GO:0007169
GO:0055085
Topology
Subcellular location
Mitochondrion inner membrane
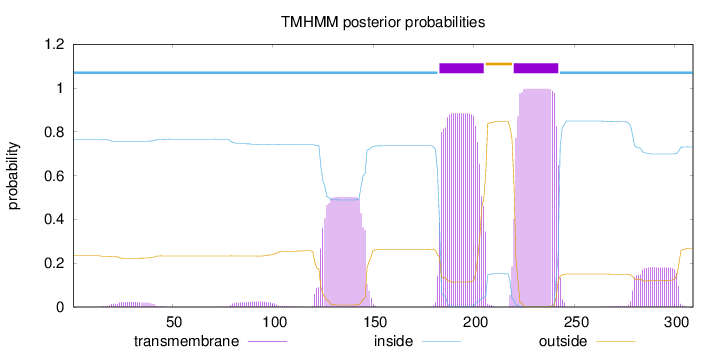
Length:
309
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
56.82948
Exp number, first 60 AAs:
0.45538
Total prob of N-in:
0.76630
inside
1 - 182
TMhelix
183 - 205
outside
206 - 219
TMhelix
220 - 242
inside
243 - 309
Population Genetic Test Statistics
Pi
7.01544
Theta
7.841508
Tajima's D
-0.795705
CLR
0.024204
CSRT
0.182040897955102
Interpretation
Uncertain
