Gene
KWMTBOMO00143
Pre Gene Modal
BGIBMGA002147
Annotation
Dynein_heavy_chain_5?_axonemal_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 2.847
Sequence
CDS
ATGCAACCGCAGTTTGAAGAGGATTTGAAAAACAATTTGGACAAATTTAGGGATGACAACGCCGAATACTGTCACGAGTATAGACACGCTGGACCGATGCAGCCAGGGTTGACCCCACGAGAAGCATCCGATAGATTGATACTTTTTCAAAATCGGTTCGACGGAATGTGGCGAAAGCTTCAGACCTACCAGAACGGTGAAGAGCTTTTTGGTCTTCCCCATACAGAATACCCTGAATTAGCCCAAATAAGAAAGGAACTGAACTTACTCCAGAAGCTGTATAAGCTTTACAACGACGTTATTGACAGGGTCAGTAGTTACTACGACATACCCTGGGGAGAAGTTAATATTGAAGAGATCAACAATGAACTGATGGAGTTTCAGAACAGATGCCGAAAGTTGCCGAAAGGTCTCAAAGAGTGGCCGGCATTCTTTGCCCTTAAGAAGACCATCGACGATTTCAACGATATGTGTCCTCTCCTTGAGCTGATGGCCAACAAAGCTATGAAGCCTCGCCATTGGCAGAGGATCATGGAGGTCACGAAATACGTTTTTGAGTTAGACAACGAGGACTTCTGCTTGAAAAATATACTAGAAGCACCCCTGCTGCAGAACAAGGAGGATATCGAGGTAAGTAGTAGAACTGAGGCTTTAAACAATCGATTCACTTATGACATCGTGCATAAGTACTAA
Protein
MQPQFEEDLKNNLDKFRDDNAEYCHEYRHAGPMQPGLTPREASDRLILFQNRFDGMWRKLQTYQNGEELFGLPHTEYPELAQIRKELNLLQKLYKLYNDVIDRVSSYYDIPWGEVNIEEINNELMEFQNRCRKLPKGLKEWPAFFALKKTIDDFNDMCPLLELMANKAMKPRHWQRIMEVTKYVFELDNEDFCLKNILEAPLLQNKEDIEVSSRTEALNNRFTYDIVHKY
Summary
Uniprot
A0A2H1VRY4
A0A2A4IX12
A0A194Q4X1
A0A194QUB4
A0A212FHI9
A0A1B6EKS6
+ More
A0A151WTR8 A0A195D4P3 A0A336LFL1 A0A3Q0J3D6 E2CA47 T1GKZ1 A0A195FIH3 A0A151J9E5 A0A158P2P5 A0A195BBY4 E2AZZ0 D6WIS3 U4U2X2 N6U4W8 F4WDX4 A0A0J7L6K6 A0A1B0GJY1 E0W0Q7 K7J7S2 A0A232F2W8 A0A154PFK8 A0A0K8ULD1 A0A336LPD9 A0A0L7QTW2 A0A1B0G5G6 A0A1A9ZNV3 A0A1A9Y7F3 A0A1B0C3G6 A0A310SAR6 A0A182T3K7 A0A1A9VBD6 A0A0M4EJY3 A0A1A9WJE8 A0A088AS19 A0A026WY55 B4N841 A0A0Q9X3M2 A0A0L0CCG7 B4M499 A0A0Q9W1A9 A0A0R3NPI5 A0A0R3NJK3 A0A0R3NQ41 B5DVH1 A0A0R3NJX4 A0A0B4K614 A0A0R3NNR0 A0A3B0KP73 Q9VH97 A0A1I8M7D6 A0A1I8PHL1 B4QWI5 A0A0R1E219 A0A0B4LH20 A0A0R1E7J6 B3M1H1 B3NZN0 A0A0B4K648 B4PVC1 T1HLR4 B4JFF8 B4K645 B4HJW0 A0A1W4W0B5 A0A1W4W0Z8 B0WIM3 A0A1J1ICX5 A0A182JFK1 A0A182QZT5 A0A084W3D4 A0A182N569 A0A182RHW0 A0A182YK77 A0A182VQN9 A0A182X1W7 A0A182VEN6 Q7PTQ4 A0A182JRZ0 A0A182HJD6 Q16UI0 A0A1S4FNK9 A0A1Q3FM85 A0A182PIC7 A0A182LCP2 A0A2S2Q235 A0A2S2PXW6 A0A182FSH4 A0A1Q3FSZ1 J9JXN3 A0A0M8ZZ88 A0A182MWN1 A0A182TQ18 A0A0N7Z9W2 A0A1B0GQ26
A0A151WTR8 A0A195D4P3 A0A336LFL1 A0A3Q0J3D6 E2CA47 T1GKZ1 A0A195FIH3 A0A151J9E5 A0A158P2P5 A0A195BBY4 E2AZZ0 D6WIS3 U4U2X2 N6U4W8 F4WDX4 A0A0J7L6K6 A0A1B0GJY1 E0W0Q7 K7J7S2 A0A232F2W8 A0A154PFK8 A0A0K8ULD1 A0A336LPD9 A0A0L7QTW2 A0A1B0G5G6 A0A1A9ZNV3 A0A1A9Y7F3 A0A1B0C3G6 A0A310SAR6 A0A182T3K7 A0A1A9VBD6 A0A0M4EJY3 A0A1A9WJE8 A0A088AS19 A0A026WY55 B4N841 A0A0Q9X3M2 A0A0L0CCG7 B4M499 A0A0Q9W1A9 A0A0R3NPI5 A0A0R3NJK3 A0A0R3NQ41 B5DVH1 A0A0R3NJX4 A0A0B4K614 A0A0R3NNR0 A0A3B0KP73 Q9VH97 A0A1I8M7D6 A0A1I8PHL1 B4QWI5 A0A0R1E219 A0A0B4LH20 A0A0R1E7J6 B3M1H1 B3NZN0 A0A0B4K648 B4PVC1 T1HLR4 B4JFF8 B4K645 B4HJW0 A0A1W4W0B5 A0A1W4W0Z8 B0WIM3 A0A1J1ICX5 A0A182JFK1 A0A182QZT5 A0A084W3D4 A0A182N569 A0A182RHW0 A0A182YK77 A0A182VQN9 A0A182X1W7 A0A182VEN6 Q7PTQ4 A0A182JRZ0 A0A182HJD6 Q16UI0 A0A1S4FNK9 A0A1Q3FM85 A0A182PIC7 A0A182LCP2 A0A2S2Q235 A0A2S2PXW6 A0A182FSH4 A0A1Q3FSZ1 J9JXN3 A0A0M8ZZ88 A0A182MWN1 A0A182TQ18 A0A0N7Z9W2 A0A1B0GQ26
Pubmed
26354079
22118469
20798317
21347285
18362917
19820115
+ More
23537049 21719571 20566863 20075255 28648823 24508170 17994087 26108605 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 17550304 24438588 25244985 12364791 14747013 17210077 17510324 20966253
23537049 21719571 20566863 20075255 28648823 24508170 17994087 26108605 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 17550304 24438588 25244985 12364791 14747013 17210077 17510324 20966253
EMBL
ODYU01003896
SOQ43222.1
NWSH01005052
PCG64477.1
KQ459463
KPJ00583.1
+ More
KQ461108 KPJ09118.1 AGBW02008503 OWR53211.1 GECZ01031268 JAS38501.1 KQ982753 KYQ51236.1 KQ976885 KYN07399.1 UFQS01004939 UFQT01004939 SSX16626.1 SSX35823.1 GL453920 EFN75202.1 CAQQ02108091 KQ981523 KYN40198.1 KQ979404 KYN21688.1 ADTU01007305 KQ976530 KYM81709.1 GL444289 EFN60993.1 KQ971321 EEZ99499.2 KB631941 ERL87387.1 APGK01049735 APGK01049736 KB741156 ENN73607.1 GL888096 EGI67606.1 LBMM01000498 KMQ98208.1 AJWK01022437 AJWK01022438 AJWK01022439 DS235862 EEB19213.1 AAZX01003355 AAZX01003361 AAZX01011149 NNAY01001113 OXU25094.1 KQ434893 KZC10625.1 GDHF01024953 JAI27361.1 UFQT01000100 SSX19932.1 KQ414736 KOC62093.1 CCAG010000321 JXJN01024951 KQ765917 OAD53781.1 CP012526 ALC45340.1 KK107069 EZA60758.1 CH964232 EDW81292.2 KRF99424.1 JRES01000608 KNC29950.1 CH940652 EDW59460.2 KRF78860.1 CM000070 KRT01085.1 KRT01086.1 KRT01083.1 KRT01088.1 EDY68486.3 KRT01084.1 AE014297 AFH06325.1 KRT01087.1 OUUW01000014 SPP88419.1 AAF54422.5 CM000364 EDX13589.1 CM000160 KRK03304.1 AHN57255.1 KRK03303.1 CH902617 EDV43262.2 CH954181 EDV49740.1 AFH06326.1 EDW96694.2 ACPB03010558 ACPB03010559 CH916369 EDV93439.1 CH933806 EDW14095.2 CH480815 EDW42844.1 DS231950 EDS28578.1 CVRI01000047 CRK98135.1 AXCN02000174 ATLV01019918 ATLV01019919 ATLV01019920 KE525285 KFB44728.1 AAAB01008794 EAA03542.6 APCN01002153 CH477621 EAT38201.1 GFDL01006315 JAV28730.1 GGMS01002633 MBY71836.1 GGMS01001007 MBY70210.1 GFDL01004377 JAV30668.1 ABLF02035929 ABLF02035940 KQ435816 KOX72633.1 AXCM01003484 AXCM01003485 GDRN01108828 JAI57160.1 AJVK01016013
KQ461108 KPJ09118.1 AGBW02008503 OWR53211.1 GECZ01031268 JAS38501.1 KQ982753 KYQ51236.1 KQ976885 KYN07399.1 UFQS01004939 UFQT01004939 SSX16626.1 SSX35823.1 GL453920 EFN75202.1 CAQQ02108091 KQ981523 KYN40198.1 KQ979404 KYN21688.1 ADTU01007305 KQ976530 KYM81709.1 GL444289 EFN60993.1 KQ971321 EEZ99499.2 KB631941 ERL87387.1 APGK01049735 APGK01049736 KB741156 ENN73607.1 GL888096 EGI67606.1 LBMM01000498 KMQ98208.1 AJWK01022437 AJWK01022438 AJWK01022439 DS235862 EEB19213.1 AAZX01003355 AAZX01003361 AAZX01011149 NNAY01001113 OXU25094.1 KQ434893 KZC10625.1 GDHF01024953 JAI27361.1 UFQT01000100 SSX19932.1 KQ414736 KOC62093.1 CCAG010000321 JXJN01024951 KQ765917 OAD53781.1 CP012526 ALC45340.1 KK107069 EZA60758.1 CH964232 EDW81292.2 KRF99424.1 JRES01000608 KNC29950.1 CH940652 EDW59460.2 KRF78860.1 CM000070 KRT01085.1 KRT01086.1 KRT01083.1 KRT01088.1 EDY68486.3 KRT01084.1 AE014297 AFH06325.1 KRT01087.1 OUUW01000014 SPP88419.1 AAF54422.5 CM000364 EDX13589.1 CM000160 KRK03304.1 AHN57255.1 KRK03303.1 CH902617 EDV43262.2 CH954181 EDV49740.1 AFH06326.1 EDW96694.2 ACPB03010558 ACPB03010559 CH916369 EDV93439.1 CH933806 EDW14095.2 CH480815 EDW42844.1 DS231950 EDS28578.1 CVRI01000047 CRK98135.1 AXCN02000174 ATLV01019918 ATLV01019919 ATLV01019920 KE525285 KFB44728.1 AAAB01008794 EAA03542.6 APCN01002153 CH477621 EAT38201.1 GFDL01006315 JAV28730.1 GGMS01002633 MBY71836.1 GGMS01001007 MBY70210.1 GFDL01004377 JAV30668.1 ABLF02035929 ABLF02035940 KQ435816 KOX72633.1 AXCM01003484 AXCM01003485 GDRN01108828 JAI57160.1 AJVK01016013
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000075809
UP000078542
+ More
UP000079169 UP000008237 UP000015102 UP000078541 UP000078492 UP000005205 UP000078540 UP000000311 UP000007266 UP000030742 UP000019118 UP000007755 UP000036403 UP000092461 UP000009046 UP000002358 UP000215335 UP000076502 UP000053825 UP000092444 UP000092445 UP000092443 UP000092460 UP000075901 UP000078200 UP000092553 UP000091820 UP000005203 UP000053097 UP000007798 UP000037069 UP000008792 UP000001819 UP000000803 UP000268350 UP000095301 UP000095300 UP000000304 UP000002282 UP000007801 UP000008711 UP000015103 UP000001070 UP000009192 UP000001292 UP000192221 UP000002320 UP000183832 UP000075880 UP000075886 UP000030765 UP000075884 UP000075900 UP000076408 UP000075920 UP000076407 UP000075903 UP000007062 UP000075881 UP000075840 UP000008820 UP000075885 UP000075882 UP000069272 UP000007819 UP000053105 UP000075883 UP000075902 UP000092462
UP000079169 UP000008237 UP000015102 UP000078541 UP000078492 UP000005205 UP000078540 UP000000311 UP000007266 UP000030742 UP000019118 UP000007755 UP000036403 UP000092461 UP000009046 UP000002358 UP000215335 UP000076502 UP000053825 UP000092444 UP000092445 UP000092443 UP000092460 UP000075901 UP000078200 UP000092553 UP000091820 UP000005203 UP000053097 UP000007798 UP000037069 UP000008792 UP000001819 UP000000803 UP000268350 UP000095301 UP000095300 UP000000304 UP000002282 UP000007801 UP000008711 UP000015103 UP000001070 UP000009192 UP000001292 UP000192221 UP000002320 UP000183832 UP000075880 UP000075886 UP000030765 UP000075884 UP000075900 UP000076408 UP000075920 UP000076407 UP000075903 UP000007062 UP000075881 UP000075840 UP000008820 UP000075885 UP000075882 UP000069272 UP000007819 UP000053105 UP000075883 UP000075902 UP000092462
PRIDE
Pfam
Interpro
IPR013602
Dynein_heavy_dom-2
+ More
IPR026983 DHC_fam
IPR013594 Dynein_heavy_dom-1
IPR041658 AAA_lid_11
IPR041228 Dynein_C
IPR035699 AAA_6
IPR004273 Dynein_heavy_D6_P-loop
IPR003593 AAA+_ATPase
IPR041466 Dynein_AAA5_ext
IPR027417 P-loop_NTPase
IPR024743 Dynein_HC_stalk
IPR024317 Dynein_heavy_chain_D4_dom
IPR035706 AAA_9
IPR041589 DNAH3_AAA_lid_1
IPR026983 DHC_fam
IPR013594 Dynein_heavy_dom-1
IPR041658 AAA_lid_11
IPR041228 Dynein_C
IPR035699 AAA_6
IPR004273 Dynein_heavy_D6_P-loop
IPR003593 AAA+_ATPase
IPR041466 Dynein_AAA5_ext
IPR027417 P-loop_NTPase
IPR024743 Dynein_HC_stalk
IPR024317 Dynein_heavy_chain_D4_dom
IPR035706 AAA_9
IPR041589 DNAH3_AAA_lid_1
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2H1VRY4
A0A2A4IX12
A0A194Q4X1
A0A194QUB4
A0A212FHI9
A0A1B6EKS6
+ More
A0A151WTR8 A0A195D4P3 A0A336LFL1 A0A3Q0J3D6 E2CA47 T1GKZ1 A0A195FIH3 A0A151J9E5 A0A158P2P5 A0A195BBY4 E2AZZ0 D6WIS3 U4U2X2 N6U4W8 F4WDX4 A0A0J7L6K6 A0A1B0GJY1 E0W0Q7 K7J7S2 A0A232F2W8 A0A154PFK8 A0A0K8ULD1 A0A336LPD9 A0A0L7QTW2 A0A1B0G5G6 A0A1A9ZNV3 A0A1A9Y7F3 A0A1B0C3G6 A0A310SAR6 A0A182T3K7 A0A1A9VBD6 A0A0M4EJY3 A0A1A9WJE8 A0A088AS19 A0A026WY55 B4N841 A0A0Q9X3M2 A0A0L0CCG7 B4M499 A0A0Q9W1A9 A0A0R3NPI5 A0A0R3NJK3 A0A0R3NQ41 B5DVH1 A0A0R3NJX4 A0A0B4K614 A0A0R3NNR0 A0A3B0KP73 Q9VH97 A0A1I8M7D6 A0A1I8PHL1 B4QWI5 A0A0R1E219 A0A0B4LH20 A0A0R1E7J6 B3M1H1 B3NZN0 A0A0B4K648 B4PVC1 T1HLR4 B4JFF8 B4K645 B4HJW0 A0A1W4W0B5 A0A1W4W0Z8 B0WIM3 A0A1J1ICX5 A0A182JFK1 A0A182QZT5 A0A084W3D4 A0A182N569 A0A182RHW0 A0A182YK77 A0A182VQN9 A0A182X1W7 A0A182VEN6 Q7PTQ4 A0A182JRZ0 A0A182HJD6 Q16UI0 A0A1S4FNK9 A0A1Q3FM85 A0A182PIC7 A0A182LCP2 A0A2S2Q235 A0A2S2PXW6 A0A182FSH4 A0A1Q3FSZ1 J9JXN3 A0A0M8ZZ88 A0A182MWN1 A0A182TQ18 A0A0N7Z9W2 A0A1B0GQ26
A0A151WTR8 A0A195D4P3 A0A336LFL1 A0A3Q0J3D6 E2CA47 T1GKZ1 A0A195FIH3 A0A151J9E5 A0A158P2P5 A0A195BBY4 E2AZZ0 D6WIS3 U4U2X2 N6U4W8 F4WDX4 A0A0J7L6K6 A0A1B0GJY1 E0W0Q7 K7J7S2 A0A232F2W8 A0A154PFK8 A0A0K8ULD1 A0A336LPD9 A0A0L7QTW2 A0A1B0G5G6 A0A1A9ZNV3 A0A1A9Y7F3 A0A1B0C3G6 A0A310SAR6 A0A182T3K7 A0A1A9VBD6 A0A0M4EJY3 A0A1A9WJE8 A0A088AS19 A0A026WY55 B4N841 A0A0Q9X3M2 A0A0L0CCG7 B4M499 A0A0Q9W1A9 A0A0R3NPI5 A0A0R3NJK3 A0A0R3NQ41 B5DVH1 A0A0R3NJX4 A0A0B4K614 A0A0R3NNR0 A0A3B0KP73 Q9VH97 A0A1I8M7D6 A0A1I8PHL1 B4QWI5 A0A0R1E219 A0A0B4LH20 A0A0R1E7J6 B3M1H1 B3NZN0 A0A0B4K648 B4PVC1 T1HLR4 B4JFF8 B4K645 B4HJW0 A0A1W4W0B5 A0A1W4W0Z8 B0WIM3 A0A1J1ICX5 A0A182JFK1 A0A182QZT5 A0A084W3D4 A0A182N569 A0A182RHW0 A0A182YK77 A0A182VQN9 A0A182X1W7 A0A182VEN6 Q7PTQ4 A0A182JRZ0 A0A182HJD6 Q16UI0 A0A1S4FNK9 A0A1Q3FM85 A0A182PIC7 A0A182LCP2 A0A2S2Q235 A0A2S2PXW6 A0A182FSH4 A0A1Q3FSZ1 J9JXN3 A0A0M8ZZ88 A0A182MWN1 A0A182TQ18 A0A0N7Z9W2 A0A1B0GQ26
PDB
5NUG
E-value=0.00737685,
Score=90
Ontologies
GO
PANTHER
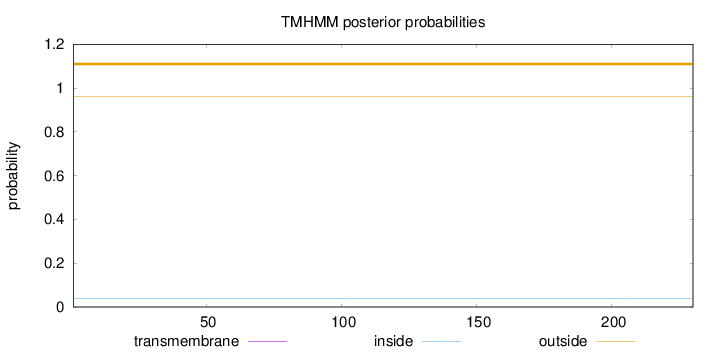
Topology
Length:
230
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03711
outside
1 - 230
Population Genetic Test Statistics
Pi
3.097505
Theta
11.599243
Tajima's D
-2.134
CLR
0
CSRT
0.00694965251737413
Interpretation
Uncertain
