Gene
KWMTBOMO00139
Pre Gene Modal
BGIBMGA002142
Annotation
PREDICTED:_arf-GAP_with_Rho-GAP_domain?_ANK_repeat_and_PH_domain-containing_protein_2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.559 Nuclear Reliability : 1.724
Sequence
CDS
ATGCTTAAACCTTGTGGAAGTCTCACTGAGGGCATATACCGTCGAGCGGGTAGCAGTTCAGTATTGTCTGAGCTGCTCGCCAGGTTCAGGAGGGACTCGTGGTCGGTCCAGTTGAGTCCCGGTCAACATTCGGAGCACGACGTCGCCGGTGTACTAAAAAGATTCTTCAGGGATCTACCAGAGTCACTTATACCTAAGGACAATCATCAGGTTCTGATCGGCGCGCTCGAAATGAAAGATGAGACGTCCCGCCACGCGGAGTACCGCCGTGTGATGTCGTCCCTATCCCTGGTCCCTCGGAACACGGCCCGGAAGTTGTTCGCTCATCTACATTTCCTGCACACGATGTCGCACGCGAATAAAATGGGCGCAGAAAATTTGGCTTCCGTTTGGGCGCCCACAATAATGCCGACCGCTTTGACGAGCAACAATTTGCAAACTGCCTGGTCGTCGAAAGAGGTGCTAGTCGTTAGAGATCTGATCGCGAACTTCGAAGCAATATGGGAGCCTACTGAAGCGGAGAAGAGACGCGAGGCGGCCGTCAGGAGAGTACTGATGAGGGTGCTGAGTAATTCAGCTCCGGCGCCGCACAGGGCCGCTGGCGACCTCAAGGCCTGGATCCACGTGAACGACAGGAGTCAGTGCTATCAAGTGGCGCTCACCCCTAACAAGACGAGCTCCGACGTATGCATAGAGCTGTGCGAGAAGGCCAAAACCGAATCGCATCTGCTGATGCTCGAAGAGGTTGTGTGTAACGAGTCGATGCGTCGTATAGTGCACATTGACGAGATCGTACTAGACGTGGTGCTCAGGTGGGGGTATTGGGACGAGGACGACAGAAAGGATAATTATTTGCTGGTGAGAGATAATAAAACGCTTCAGGATATGGATTCGTTGAGACAGACCACGTCGCTTGTGTGCGGCGAAATGAGGTTCGCAAATGAATCGATGAAGACATTCAAACAGCACATGTTCGAGCTGCAGAATAGATGCTTGTGTTACTTCAAAGATAAACAGGGTTCGCAGAAAATCGAAGAATGGAACGTCAAAGATATCCTATGGTACATCGGCCACGAACCGAAACGTAATCCACAAACCCGTTGGGCGATCACCTTTATACCGAGGAATAAAGAAAAAAGGAGCAAAGAGAAACCGTGGTTCGGTAAGACAATAGCCGGCGCGGTAACGGAAGATCAACTAAAGTGGATGTCGGCACTGATGTTCGCCGAGCACACGAGCATACTACCAACGCCTCGACTGGTTATAACATAG
Protein
MLKPCGSLTEGIYRRAGSSSVLSELLARFRRDSWSVQLSPGQHSEHDVAGVLKRFFRDLPESLIPKDNHQVLIGALEMKDETSRHAEYRRVMSSLSLVPRNTARKLFAHLHFLHTMSHANKMGAENLASVWAPTIMPTALTSNNLQTAWSSKEVLVVRDLIANFEAIWEPTEAEKRREAAVRRVLMRVLSNSAPAPHRAAGDLKAWIHVNDRSQCYQVALTPNKTSSDVCIELCEKAKTESHLLMLEEVVCNESMRRIVHIDEIVLDVVLRWGYWDEDDRKDNYLLVRDNKTLQDMDSLRQTTSLVCGEMRFANESMKTFKQHMFELQNRCLCYFKDKQGSQKIEEWNVKDILWYIGHEPKRNPQTRWAITFIPRNKEKRSKEKPWFGKTIAGAVTEDQLKWMSALMFAEHTSILPTPRLVIT
Summary
Similarity
Belongs to the GST superfamily.
Uniprot
A0A212F9S2
A0A2H1WGX4
A0A194QZU8
A0A194QAY3
A0A2Z5U7E1
A0A2J7QZA1
+ More
A0A1B6MRT4 A0A1B6LEB6 A0A1B6H8G0 A0A1B6IHX6 A0A154PJR1 A0A1B6EQL3 A0A1B6MF42 A0A1B6KCU6 A0A1S4ESY4 A0A0C9R1W2 A0A026WSZ2 A0A3L8DH35 E2B7Q3 A0A195EGC3 A0A151WR18 F4WY92 A0A195AX45 A0A195FEN2 A0A195CBK1 A0A158NHS5 E2AZR0 A0A0J7LBB0 A0A0V0GB74 A0A224XQD9 A0A0L7R9F2 A0A1W4XRT0 E9JB82 A0A0C9PZ76 A0A0P4VHG3 A0A2A3E2V2 A0A088AI97 A0A1Y1L1F6 E0VT06 A0A069DXX1 A0A2S2NUS8 A0A2H8TH91 J9K1U7 A0A2S2QZW1 A0A2R7WM86 D6WE41 A0A0T6B4U2 A0A2P8YAZ8 A0A0N1ITB7 U4UVV5 A0A310SEM9 N6TD68 A0A1B6EDQ8 A0A0C9REQ4 A0A1B0CEA4 A0A1J1HH09 A0A226CXT8 A0A336KS94 U5EDU6 T1PPN4 A0A034VFJ1 A0A034VI84 A0A0P6IVV9 Q17EB0 A0A0K8TX96 A0A0K8VJ81 A0A1I8M272 A0A0L0BT30 W8AB86 A0A182K8D2 A0A1I8Q100 A0A2M3Z3V4 A0A1S4F5Y7 A0A182H3M4 A0A3B0JDC2 A0A2M3ZGM3 A0A182KY66 A0A3B0JZF3 A0A3B0JRL9 A0A0K2U9E3 Q8T3K2 A0A0P9AHP2 B3N0B3 A0A1W4VX72 A0A182FJX9 A0A1W4VJ66 A0A2M4BB80 B4IEV9 A0A0Q5TJ75 B3NWA5 Q0KHR5 A0A0R1EBN8 A0A2M4A3V9 Q9VX92 B4Q244 A0A2M4A3W7 A0A2M4BAU3 A0A0R1EID4 A0A182PTU5
A0A1B6MRT4 A0A1B6LEB6 A0A1B6H8G0 A0A1B6IHX6 A0A154PJR1 A0A1B6EQL3 A0A1B6MF42 A0A1B6KCU6 A0A1S4ESY4 A0A0C9R1W2 A0A026WSZ2 A0A3L8DH35 E2B7Q3 A0A195EGC3 A0A151WR18 F4WY92 A0A195AX45 A0A195FEN2 A0A195CBK1 A0A158NHS5 E2AZR0 A0A0J7LBB0 A0A0V0GB74 A0A224XQD9 A0A0L7R9F2 A0A1W4XRT0 E9JB82 A0A0C9PZ76 A0A0P4VHG3 A0A2A3E2V2 A0A088AI97 A0A1Y1L1F6 E0VT06 A0A069DXX1 A0A2S2NUS8 A0A2H8TH91 J9K1U7 A0A2S2QZW1 A0A2R7WM86 D6WE41 A0A0T6B4U2 A0A2P8YAZ8 A0A0N1ITB7 U4UVV5 A0A310SEM9 N6TD68 A0A1B6EDQ8 A0A0C9REQ4 A0A1B0CEA4 A0A1J1HH09 A0A226CXT8 A0A336KS94 U5EDU6 T1PPN4 A0A034VFJ1 A0A034VI84 A0A0P6IVV9 Q17EB0 A0A0K8TX96 A0A0K8VJ81 A0A1I8M272 A0A0L0BT30 W8AB86 A0A182K8D2 A0A1I8Q100 A0A2M3Z3V4 A0A1S4F5Y7 A0A182H3M4 A0A3B0JDC2 A0A2M3ZGM3 A0A182KY66 A0A3B0JZF3 A0A3B0JRL9 A0A0K2U9E3 Q8T3K2 A0A0P9AHP2 B3N0B3 A0A1W4VX72 A0A182FJX9 A0A1W4VJ66 A0A2M4BB80 B4IEV9 A0A0Q5TJ75 B3NWA5 Q0KHR5 A0A0R1EBN8 A0A2M4A3V9 Q9VX92 B4Q244 A0A2M4A3W7 A0A2M4BAU3 A0A0R1EID4 A0A182PTU5
Pubmed
22118469
26354079
24508170
30249741
20798317
21719571
+ More
21347285 21282665 27129103 28004739 20566863 26334808 18362917 19820115 29403074 23537049 25348373 26999592 17510324 25315136 26108605 24495485 26483478 20966253 17994087 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304
21347285 21282665 27129103 28004739 20566863 26334808 18362917 19820115 29403074 23537049 25348373 26999592 17510324 25315136 26108605 24495485 26483478 20966253 17994087 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304
EMBL
AGBW02009564
OWR50495.1
ODYU01008593
SOQ52318.1
KQ461108
KPJ09116.1
+ More
KQ459463 KPJ00581.1 AP017515 BBB06853.1 NEVH01009072 PNF33895.1 GEBQ01001382 JAT38595.1 GEBQ01017961 JAT22016.1 GECU01036747 JAS70959.1 GECU01021177 JAS86529.1 KQ434938 KZC12099.1 GECZ01029535 GECZ01013369 GECZ01009669 GECZ01000977 JAS40234.1 JAS56400.1 JAS60100.1 JAS68792.1 GEBQ01005399 JAT34578.1 GEBQ01030698 JAT09279.1 GBYB01006812 JAG76579.1 KK107109 EZA59170.1 QOIP01000008 RLU19239.1 GL446201 EFN88266.1 KQ978957 KYN27288.1 KQ982813 KYQ50253.1 GL888439 EGI60849.1 KQ976725 KYM76617.1 KQ981636 KYN38858.1 KQ978023 KYM98075.1 ADTU01016037 GL444277 EFN61039.1 LBMM01000040 KMR05197.1 GECL01000862 JAP05262.1 GFTR01006063 JAW10363.1 KQ414627 KOC67459.1 GL770938 EFZ09896.1 GBYB01006808 JAG76575.1 GDKW01003495 JAI53100.1 KZ288462 PBC25469.1 GEZM01070890 JAV66230.1 AAZO01005192 DS235759 EEB16512.1 GBGD01000208 JAC88681.1 GGMR01008285 MBY20904.1 GFXV01001585 MBW13390.1 ABLF02023675 ABLF02023676 ABLF02023677 GGMS01014062 MBY83265.1 KK855086 PTY20743.1 KQ971318 EFA00372.1 LJIG01009871 KRT82213.1 PYGN01000743 PSN41421.1 KQ435828 KOX71878.1 KB632385 ERL94330.1 KQ769940 OAD52745.1 APGK01042753 KB741007 ENN75648.1 GEDC01001222 JAS36076.1 GBYB01006810 JAG76577.1 AJWK01008702 AJWK01008703 AJWK01008704 AJWK01008705 AJWK01008706 AJWK01008707 CVRI01000004 CRK87304.1 LNIX01000048 OXA38142.1 UFQS01000919 UFQT01000919 SSX07664.1 SSX28002.1 GANO01004593 JAB55278.1 KA650023 AFP64652.1 GAKP01017718 JAC41234.1 GAKP01017709 JAC41243.1 GDUN01000807 JAN95112.1 CH477284 EAT44768.1 GDHF01033416 JAI18898.1 GDHF01013392 JAI38922.1 JRES01001383 KNC23235.1 GAMC01021445 JAB85110.1 GGFM01002439 MBW23190.1 JXUM01024585 JXUM01024586 JXUM01024587 JXUM01024588 OUUW01000003 SPP78112.1 GGFM01006945 MBW27696.1 SPP78111.1 SPP78110.1 HACA01016970 CDW34331.1 AY094969 AAM11322.1 CH902640 KPU77360.1 EDV38317.1 KPU77361.1 GGFJ01001100 MBW50241.1 CH480832 EDW46213.1 CH954180 KQS29974.1 EDV46444.1 AE014298 ABI30987.1 CM000162 KRK06827.1 GGFK01002172 MBW35493.1 AAF48686.3 EDX02552.2 GGFK01002124 MBW35445.1 GGFJ01000787 MBW49928.1 KRK06826.1
KQ459463 KPJ00581.1 AP017515 BBB06853.1 NEVH01009072 PNF33895.1 GEBQ01001382 JAT38595.1 GEBQ01017961 JAT22016.1 GECU01036747 JAS70959.1 GECU01021177 JAS86529.1 KQ434938 KZC12099.1 GECZ01029535 GECZ01013369 GECZ01009669 GECZ01000977 JAS40234.1 JAS56400.1 JAS60100.1 JAS68792.1 GEBQ01005399 JAT34578.1 GEBQ01030698 JAT09279.1 GBYB01006812 JAG76579.1 KK107109 EZA59170.1 QOIP01000008 RLU19239.1 GL446201 EFN88266.1 KQ978957 KYN27288.1 KQ982813 KYQ50253.1 GL888439 EGI60849.1 KQ976725 KYM76617.1 KQ981636 KYN38858.1 KQ978023 KYM98075.1 ADTU01016037 GL444277 EFN61039.1 LBMM01000040 KMR05197.1 GECL01000862 JAP05262.1 GFTR01006063 JAW10363.1 KQ414627 KOC67459.1 GL770938 EFZ09896.1 GBYB01006808 JAG76575.1 GDKW01003495 JAI53100.1 KZ288462 PBC25469.1 GEZM01070890 JAV66230.1 AAZO01005192 DS235759 EEB16512.1 GBGD01000208 JAC88681.1 GGMR01008285 MBY20904.1 GFXV01001585 MBW13390.1 ABLF02023675 ABLF02023676 ABLF02023677 GGMS01014062 MBY83265.1 KK855086 PTY20743.1 KQ971318 EFA00372.1 LJIG01009871 KRT82213.1 PYGN01000743 PSN41421.1 KQ435828 KOX71878.1 KB632385 ERL94330.1 KQ769940 OAD52745.1 APGK01042753 KB741007 ENN75648.1 GEDC01001222 JAS36076.1 GBYB01006810 JAG76577.1 AJWK01008702 AJWK01008703 AJWK01008704 AJWK01008705 AJWK01008706 AJWK01008707 CVRI01000004 CRK87304.1 LNIX01000048 OXA38142.1 UFQS01000919 UFQT01000919 SSX07664.1 SSX28002.1 GANO01004593 JAB55278.1 KA650023 AFP64652.1 GAKP01017718 JAC41234.1 GAKP01017709 JAC41243.1 GDUN01000807 JAN95112.1 CH477284 EAT44768.1 GDHF01033416 JAI18898.1 GDHF01013392 JAI38922.1 JRES01001383 KNC23235.1 GAMC01021445 JAB85110.1 GGFM01002439 MBW23190.1 JXUM01024585 JXUM01024586 JXUM01024587 JXUM01024588 OUUW01000003 SPP78112.1 GGFM01006945 MBW27696.1 SPP78111.1 SPP78110.1 HACA01016970 CDW34331.1 AY094969 AAM11322.1 CH902640 KPU77360.1 EDV38317.1 KPU77361.1 GGFJ01001100 MBW50241.1 CH480832 EDW46213.1 CH954180 KQS29974.1 EDV46444.1 AE014298 ABI30987.1 CM000162 KRK06827.1 GGFK01002172 MBW35493.1 AAF48686.3 EDX02552.2 GGFK01002124 MBW35445.1 GGFJ01000787 MBW49928.1 KRK06826.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000235965
UP000076502
UP000079169
+ More
UP000053097 UP000279307 UP000008237 UP000078492 UP000075809 UP000007755 UP000078540 UP000078541 UP000078542 UP000005205 UP000000311 UP000036403 UP000053825 UP000192223 UP000242457 UP000005203 UP000009046 UP000007819 UP000007266 UP000245037 UP000053105 UP000030742 UP000019118 UP000092461 UP000183832 UP000198287 UP000008820 UP000095301 UP000037069 UP000075881 UP000095300 UP000069940 UP000268350 UP000075882 UP000007801 UP000192221 UP000069272 UP000001292 UP000008711 UP000000803 UP000002282 UP000075885
UP000053097 UP000279307 UP000008237 UP000078492 UP000075809 UP000007755 UP000078540 UP000078541 UP000078542 UP000005205 UP000000311 UP000036403 UP000053825 UP000192223 UP000242457 UP000005203 UP000009046 UP000007819 UP000007266 UP000245037 UP000053105 UP000030742 UP000019118 UP000092461 UP000183832 UP000198287 UP000008820 UP000095301 UP000037069 UP000075881 UP000095300 UP000069940 UP000268350 UP000075882 UP000007801 UP000192221 UP000069272 UP000001292 UP000008711 UP000000803 UP000002282 UP000075885
Interpro
IPR000198
RhoGAP_dom
+ More
IPR008936 Rho_GTPase_activation_prot
IPR001849 PH_domain
IPR011993 PH-like_dom_sf
IPR029071 Ubiquitin-like_domsf
IPR037858 RhoGAP_ARAP
IPR000159 RA_dom
IPR005442 GST_omega
IPR036282 Glutathione-S-Trfase_C_sf
IPR036249 Thioredoxin-like_sf
IPR040079 Glutathione_S-Trfase
IPR010987 Glutathione-S-Trfase_C-like
IPR004045 Glutathione_S-Trfase_N
IPR013087 Znf_C2H2_type
IPR001478 PDZ
IPR036034 PDZ_sf
IPR008936 Rho_GTPase_activation_prot
IPR001849 PH_domain
IPR011993 PH-like_dom_sf
IPR029071 Ubiquitin-like_domsf
IPR037858 RhoGAP_ARAP
IPR000159 RA_dom
IPR005442 GST_omega
IPR036282 Glutathione-S-Trfase_C_sf
IPR036249 Thioredoxin-like_sf
IPR040079 Glutathione_S-Trfase
IPR010987 Glutathione-S-Trfase_C-like
IPR004045 Glutathione_S-Trfase_N
IPR013087 Znf_C2H2_type
IPR001478 PDZ
IPR036034 PDZ_sf
SUPFAM
Gene 3D
ProteinModelPortal
A0A212F9S2
A0A2H1WGX4
A0A194QZU8
A0A194QAY3
A0A2Z5U7E1
A0A2J7QZA1
+ More
A0A1B6MRT4 A0A1B6LEB6 A0A1B6H8G0 A0A1B6IHX6 A0A154PJR1 A0A1B6EQL3 A0A1B6MF42 A0A1B6KCU6 A0A1S4ESY4 A0A0C9R1W2 A0A026WSZ2 A0A3L8DH35 E2B7Q3 A0A195EGC3 A0A151WR18 F4WY92 A0A195AX45 A0A195FEN2 A0A195CBK1 A0A158NHS5 E2AZR0 A0A0J7LBB0 A0A0V0GB74 A0A224XQD9 A0A0L7R9F2 A0A1W4XRT0 E9JB82 A0A0C9PZ76 A0A0P4VHG3 A0A2A3E2V2 A0A088AI97 A0A1Y1L1F6 E0VT06 A0A069DXX1 A0A2S2NUS8 A0A2H8TH91 J9K1U7 A0A2S2QZW1 A0A2R7WM86 D6WE41 A0A0T6B4U2 A0A2P8YAZ8 A0A0N1ITB7 U4UVV5 A0A310SEM9 N6TD68 A0A1B6EDQ8 A0A0C9REQ4 A0A1B0CEA4 A0A1J1HH09 A0A226CXT8 A0A336KS94 U5EDU6 T1PPN4 A0A034VFJ1 A0A034VI84 A0A0P6IVV9 Q17EB0 A0A0K8TX96 A0A0K8VJ81 A0A1I8M272 A0A0L0BT30 W8AB86 A0A182K8D2 A0A1I8Q100 A0A2M3Z3V4 A0A1S4F5Y7 A0A182H3M4 A0A3B0JDC2 A0A2M3ZGM3 A0A182KY66 A0A3B0JZF3 A0A3B0JRL9 A0A0K2U9E3 Q8T3K2 A0A0P9AHP2 B3N0B3 A0A1W4VX72 A0A182FJX9 A0A1W4VJ66 A0A2M4BB80 B4IEV9 A0A0Q5TJ75 B3NWA5 Q0KHR5 A0A0R1EBN8 A0A2M4A3V9 Q9VX92 B4Q244 A0A2M4A3W7 A0A2M4BAU3 A0A0R1EID4 A0A182PTU5
A0A1B6MRT4 A0A1B6LEB6 A0A1B6H8G0 A0A1B6IHX6 A0A154PJR1 A0A1B6EQL3 A0A1B6MF42 A0A1B6KCU6 A0A1S4ESY4 A0A0C9R1W2 A0A026WSZ2 A0A3L8DH35 E2B7Q3 A0A195EGC3 A0A151WR18 F4WY92 A0A195AX45 A0A195FEN2 A0A195CBK1 A0A158NHS5 E2AZR0 A0A0J7LBB0 A0A0V0GB74 A0A224XQD9 A0A0L7R9F2 A0A1W4XRT0 E9JB82 A0A0C9PZ76 A0A0P4VHG3 A0A2A3E2V2 A0A088AI97 A0A1Y1L1F6 E0VT06 A0A069DXX1 A0A2S2NUS8 A0A2H8TH91 J9K1U7 A0A2S2QZW1 A0A2R7WM86 D6WE41 A0A0T6B4U2 A0A2P8YAZ8 A0A0N1ITB7 U4UVV5 A0A310SEM9 N6TD68 A0A1B6EDQ8 A0A0C9REQ4 A0A1B0CEA4 A0A1J1HH09 A0A226CXT8 A0A336KS94 U5EDU6 T1PPN4 A0A034VFJ1 A0A034VI84 A0A0P6IVV9 Q17EB0 A0A0K8TX96 A0A0K8VJ81 A0A1I8M272 A0A0L0BT30 W8AB86 A0A182K8D2 A0A1I8Q100 A0A2M3Z3V4 A0A1S4F5Y7 A0A182H3M4 A0A3B0JDC2 A0A2M3ZGM3 A0A182KY66 A0A3B0JZF3 A0A3B0JRL9 A0A0K2U9E3 Q8T3K2 A0A0P9AHP2 B3N0B3 A0A1W4VX72 A0A182FJX9 A0A1W4VJ66 A0A2M4BB80 B4IEV9 A0A0Q5TJ75 B3NWA5 Q0KHR5 A0A0R1EBN8 A0A2M4A3V9 Q9VX92 B4Q244 A0A2M4A3W7 A0A2M4BAU3 A0A0R1EID4 A0A182PTU5
PDB
5JCP
E-value=8.72505e-17,
Score=213
Ontologies
GO
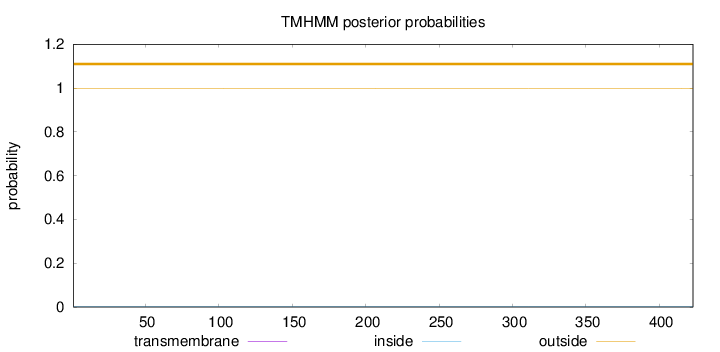
Topology
Length:
423
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00427
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00217
outside
1 - 423
Population Genetic Test Statistics
Pi
24.042402
Theta
20.050466
Tajima's D
-1.491737
CLR
0.210481
CSRT
0.0579471026448678
Interpretation
Uncertain
