Pre Gene Modal
BGIBMGA002008
Annotation
PREDICTED:_protein_phosphatase_1_regulatory_subunit_12C_isoform_X6_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.312
Sequence
CDS
ATGACGGATAACAGGTCGTCGTCAGCGTTGTTTAAGCGCGCGGAGCAACTCAAGAGATGGGAAGAGTCCGAGACAAATAAACAGTCGCCGACGCCGAAGCGCGCCTCTAGGATTCAGTTTTCCGCGGGGATTGTTTTCTTGGCTGCGTGTACGTCCGGCGACAAAGATGAAGTACAACGCCTGCTGAAGATGGGTGCCGATATCAACACGGCTAACGTGGACGGACTCACAGCTTTACATCAGGCGTGTATTGATGACAACTTGGATATGGTGGAGTTTTTAGTAACTCATGGTGCGGATGTCAACCGCGGCGACAACGAAGGCTGGACTCCACTGCACGCTACTGCTTCTTGTGGCTTCATCAGTATAGCAAGGTATCTCCTTGAGAGCGGTGCGGACGTGGCCGCGGTCAACTATGATGGCGAGTTGCCTGTTGATATCGCCGAAAGCGATGCAATGGCTGACTTGCTACAGAAAGTAATTGACGAGAAAGGCGTTGATTGTGAGAAGTCAAGGAATGCAGAAGTTGATACGCTGATGAGTGACGCCAGAGAATGGGCATTGCACGGCTACGTTGAAGTTCGAGACTTGAAAACGGGAGGCACGCCGCTCCACGTTGCCGCTGCTAAAGGATATATCGACGTGGCCAAAACTCTACTGGAGGATTGTAATGCGGATCCTGACTGCGTTGACTACGAGGGCTGGACACCGTTGCACGCGGCCGCATTGTGGGGACAAAAGGAGGCGGCAGCATTACTACTCAAATTCGGAGCTGATCCACATCTCAAAAACTATTCGGGACAGACCTGCTTAGATCTGGTGGATCCGAGCATCTCGTCATGGCTGGAGGAGGCAGCCCTGAAGGCTCCCCGTCGCGTCAATAACAACAAACGGAAACTTAGCCCTCCGAGGCACGAAGTCAAAAGAATCGACATCGAAATCAAGGGACAAGCAGAGAAAATTGTCAACGACAAACCGCCGCCGCCCGAGATCATTGCTAAGAAGGCCGAGAAGCCACCAAAGGGCCGGGCTCCGTCCCCCGAAAGCACAGAGATGGCGGAACAACAGGCACAGAACGATGAGGCGCCCTCGTGGCGTAGATCTGCCTCGTTTAGGAACAGGCAACAAGACTCAGCCAAAGAAAACAAACCGGCCGTGAACAATGTCGACGACACGATACTGAGGAGGACGCACAGCTTCGAAAACGACAAGACAATAAGTCCAGCGGAAAGAAGGGATAAAGAAAATAACGCGAGAATTGCGCTGAGCGCGACAAGCAACGCCAACAACGCGACAACGAACTCCCAGGCGCCGGTCAGGAGATCGTTCGTGCCCCCGGTCCGCGACGAGGAGAGCGAAACGCAAAGGAAGGCGCACGCGAAACGGGTCCGCGAGACTCGACGTTCCACCCAGGGCGTTACCCTCGATGAGATTAAATCAGCCGAACAGCTTGTCAAAAAGAAATCCAGCAACGGCACGACGGAGAGTTCCACTCCGACTAACGTCAAAAAGGATGAAACTCCAACTCCGGCGACCACAGACAACGGATCGTTCGAGCTGGAGGACGCCGGTAGAGCGTCGCCTCGATCGGGGGCAGCCGAAGCGACGGTCACCCTGCCGCTGCGTCGACCTCCGTCAGCATCGACGAACGCCACGACGGCCGCCGCAACTAAAGAAGACAACCGCACCGAAGTTGAAGAGGAGAAAGACAGAGAAAGCAGTACGGAGAGCAAGTCGAGTAACGCCTCGCCTGCCAGCACGCAGGCGGCGCTCAATGTCATACAGCGTCGACGCAGGCCGAAGAGACGGTCGACCGGCGTCGGACATGTGGATATGGACGTAAGTGACACTAGATTCACCGAACTAATAGGTGAGCTCACGGGGCTCAAGCCGGGAGTGTTGCTAAAACTAGCTCTAGCAAAATGCAGTGCTTCGTAG
Protein
MTDNRSSSALFKRAEQLKRWEESETNKQSPTPKRASRIQFSAGIVFLAACTSGDKDEVQRLLKMGADINTANVDGLTALHQACIDDNLDMVEFLVTHGADVNRGDNEGWTPLHATASCGFISIARYLLESGADVAAVNYDGELPVDIAESDAMADLLQKVIDEKGVDCEKSRNAEVDTLMSDAREWALHGYVEVRDLKTGGTPLHVAAAKGYIDVAKTLLEDCNADPDCVDYEGWTPLHAAALWGQKEAAALLLKFGADPHLKNYSGQTCLDLVDPSISSWLEEAALKAPRRVNNNKRKLSPPRHEVKRIDIEIKGQAEKIVNDKPPPPEIIAKKAEKPPKGRAPSPESTEMAEQQAQNDEAPSWRRSASFRNRQQDSAKENKPAVNNVDDTILRRTHSFENDKTISPAERRDKENNARIALSATSNANNATTNSQAPVRRSFVPPVRDEESETQRKAHAKRVRETRRSTQGVTLDEIKSAEQLVKKKSSNGTTESSTPTNVKKDETPTPATTDNGSFELEDAGRASPRSGAAEATVTLPLRRPPSASTNATTAAATKEDNRTEVEEEKDRESSTESKSSNASPASTQAALNVIQRRRRPKRRSTGVGHVDMDVSDTRFTELIGELTGLKPGVLLKLALAKCSAS
Summary
Uniprot
H9IXM5
A0A2A4K251
A0A2A4K1R9
A0A2H1WRT9
A0A1Y1JXV5
A0A1Y1JWP9
+ More
A0A1Y1K1F1 A0A1Y1JTJ8 T1PEE5 A0A0A1XK87 T1PEI7 T1PG43 T1PLK6 E0W0P8 A0A0A1WF26 A0A034V805 A0A034V5X0 A0A1I8NAM0 A0A1L8DMU0 A0A1Q3FTQ1 A0A1Q3FTS8 A0A1Q3FTL7 A0A1Q3FTM3 A0A1Q3FTL2 A0A146KTI9 A0A034V709 A0A034V3N2 A0A1W4UTM9 A0A2J7QEF9 A0A2J7QED8 A0A0Q5U557 A0A0J9RXH5 A0A1W4UTX3 Q86R99 A8JNT5 A0A1W4UG54 A0A0Q5UH57 A0A0J9RW24 A0A0Q5U8Y8 A0A1W4UTW8 A0A0R3P353 A0A0J9RW22 A0A0R3P811 A0A0R3P3C7 A0A1W4UGQ2 M9PCQ3 Q8MV37 A0A0P8XVK7 A0A0Q9WQJ7 A0A0Q9XBS6 B4L081 A0A0Q9XBJ6 A0A084WA83 A0A0P8XVL6 A0A0Q9WI93 A0A0P8XVM6 A0A1W4UGP7 A0A1W4UVB6 A0A0J9RWE5 A0A2S2QF17 B4HJ07 B3NDR1 Q8IQN4 Q8MT32 K7J9N7 A0A0M4ECR6 A0A0C9RGS4 A0A1B6JXK3 A0A146KR34 A0A1B6IZ70 A0A1B6GT14 A0A146M082
A0A1Y1K1F1 A0A1Y1JTJ8 T1PEE5 A0A0A1XK87 T1PEI7 T1PG43 T1PLK6 E0W0P8 A0A0A1WF26 A0A034V805 A0A034V5X0 A0A1I8NAM0 A0A1L8DMU0 A0A1Q3FTQ1 A0A1Q3FTS8 A0A1Q3FTL7 A0A1Q3FTM3 A0A1Q3FTL2 A0A146KTI9 A0A034V709 A0A034V3N2 A0A1W4UTM9 A0A2J7QEF9 A0A2J7QED8 A0A0Q5U557 A0A0J9RXH5 A0A1W4UTX3 Q86R99 A8JNT5 A0A1W4UG54 A0A0Q5UH57 A0A0J9RW24 A0A0Q5U8Y8 A0A1W4UTW8 A0A0R3P353 A0A0J9RW22 A0A0R3P811 A0A0R3P3C7 A0A1W4UGQ2 M9PCQ3 Q8MV37 A0A0P8XVK7 A0A0Q9WQJ7 A0A0Q9XBS6 B4L081 A0A0Q9XBJ6 A0A084WA83 A0A0P8XVL6 A0A0Q9WI93 A0A0P8XVM6 A0A1W4UGP7 A0A1W4UVB6 A0A0J9RWE5 A0A2S2QF17 B4HJ07 B3NDR1 Q8IQN4 Q8MT32 K7J9N7 A0A0M4ECR6 A0A0C9RGS4 A0A1B6JXK3 A0A146KR34 A0A1B6IZ70 A0A1B6GT14 A0A146M082
Pubmed
EMBL
BABH01015935
BABH01015936
BABH01015937
BABH01015938
NWSH01000275
PCG77843.1
+ More
PCG77844.1 ODYU01010520 SOQ55682.1 GEZM01101478 JAV52640.1 GEZM01101477 JAV52641.1 GEZM01101481 JAV52637.1 GEZM01101483 JAV52634.1 KA647111 AFP61740.1 GBXI01002871 JAD11421.1 KA647109 AFP61738.1 KA647110 AFP61739.1 KA649656 AFP64285.1 DS235862 EEB19204.1 GBXI01016683 JAC97608.1 GAKP01021052 JAC37900.1 GAKP01021048 JAC37904.1 GFDF01006315 JAV07769.1 GFDL01004098 JAV30947.1 GFDL01004097 JAV30948.1 GFDL01004096 JAV30949.1 GFDL01004095 JAV30950.1 GFDL01004094 JAV30951.1 GDHC01020147 JAP98481.1 GAKP01021045 JAC37907.1 GAKP01021051 JAC37901.1 NEVH01015312 PNF26966.1 PNF26952.1 CH954178 KQS44168.1 CM002912 KMY99924.1 AY048676 AAL06602.1 AE014296 ABW08547.1 KQS44163.1 KMY99916.1 KQS44164.1 CH379069 KRT07618.1 KMY99903.1 KRT07622.1 KRT07628.1 AGB94612.1 AF500094 AAM74143.1 AAN11759.2 CH902618 KPU78742.1 CH940647 KRF84407.1 CH933809 KRG05916.1 EDW18027.2 KRG05910.1 ATLV01022050 ATLV01022051 ATLV01022052 KE525327 KFB47127.1 KPU78751.1 KRF84413.1 KPU78760.1 KMY99907.1 KMY99909.1 GGMS01007121 MBY76324.1 CH480815 EDW41663.1 EDV52194.2 KQS44178.1 BT133053 AAN11758.2 AEX93139.1 AY118409 AAM48438.1 AAZX01002709 AAZX01008474 CP012525 ALC43258.1 GBYB01007555 JAG77322.1 GECU01003781 JAT03926.1 GDHC01020912 JAP97716.1 GECU01015566 JAS92140.1 GECZ01004185 JAS65584.1 GDHC01006452 JAQ12177.1
PCG77844.1 ODYU01010520 SOQ55682.1 GEZM01101478 JAV52640.1 GEZM01101477 JAV52641.1 GEZM01101481 JAV52637.1 GEZM01101483 JAV52634.1 KA647111 AFP61740.1 GBXI01002871 JAD11421.1 KA647109 AFP61738.1 KA647110 AFP61739.1 KA649656 AFP64285.1 DS235862 EEB19204.1 GBXI01016683 JAC97608.1 GAKP01021052 JAC37900.1 GAKP01021048 JAC37904.1 GFDF01006315 JAV07769.1 GFDL01004098 JAV30947.1 GFDL01004097 JAV30948.1 GFDL01004096 JAV30949.1 GFDL01004095 JAV30950.1 GFDL01004094 JAV30951.1 GDHC01020147 JAP98481.1 GAKP01021045 JAC37907.1 GAKP01021051 JAC37901.1 NEVH01015312 PNF26966.1 PNF26952.1 CH954178 KQS44168.1 CM002912 KMY99924.1 AY048676 AAL06602.1 AE014296 ABW08547.1 KQS44163.1 KMY99916.1 KQS44164.1 CH379069 KRT07618.1 KMY99903.1 KRT07622.1 KRT07628.1 AGB94612.1 AF500094 AAM74143.1 AAN11759.2 CH902618 KPU78742.1 CH940647 KRF84407.1 CH933809 KRG05916.1 EDW18027.2 KRG05910.1 ATLV01022050 ATLV01022051 ATLV01022052 KE525327 KFB47127.1 KPU78751.1 KRF84413.1 KPU78760.1 KMY99907.1 KMY99909.1 GGMS01007121 MBY76324.1 CH480815 EDW41663.1 EDV52194.2 KQS44178.1 BT133053 AAN11758.2 AEX93139.1 AY118409 AAM48438.1 AAZX01002709 AAZX01008474 CP012525 ALC43258.1 GBYB01007555 JAG77322.1 GECU01003781 JAT03926.1 GDHC01020912 JAP97716.1 GECU01015566 JAS92140.1 GECZ01004185 JAS65584.1 GDHC01006452 JAQ12177.1
Proteomes
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
CDD
ProteinModelPortal
H9IXM5
A0A2A4K251
A0A2A4K1R9
A0A2H1WRT9
A0A1Y1JXV5
A0A1Y1JWP9
+ More
A0A1Y1K1F1 A0A1Y1JTJ8 T1PEE5 A0A0A1XK87 T1PEI7 T1PG43 T1PLK6 E0W0P8 A0A0A1WF26 A0A034V805 A0A034V5X0 A0A1I8NAM0 A0A1L8DMU0 A0A1Q3FTQ1 A0A1Q3FTS8 A0A1Q3FTL7 A0A1Q3FTM3 A0A1Q3FTL2 A0A146KTI9 A0A034V709 A0A034V3N2 A0A1W4UTM9 A0A2J7QEF9 A0A2J7QED8 A0A0Q5U557 A0A0J9RXH5 A0A1W4UTX3 Q86R99 A8JNT5 A0A1W4UG54 A0A0Q5UH57 A0A0J9RW24 A0A0Q5U8Y8 A0A1W4UTW8 A0A0R3P353 A0A0J9RW22 A0A0R3P811 A0A0R3P3C7 A0A1W4UGQ2 M9PCQ3 Q8MV37 A0A0P8XVK7 A0A0Q9WQJ7 A0A0Q9XBS6 B4L081 A0A0Q9XBJ6 A0A084WA83 A0A0P8XVL6 A0A0Q9WI93 A0A0P8XVM6 A0A1W4UGP7 A0A1W4UVB6 A0A0J9RWE5 A0A2S2QF17 B4HJ07 B3NDR1 Q8IQN4 Q8MT32 K7J9N7 A0A0M4ECR6 A0A0C9RGS4 A0A1B6JXK3 A0A146KR34 A0A1B6IZ70 A0A1B6GT14 A0A146M082
A0A1Y1K1F1 A0A1Y1JTJ8 T1PEE5 A0A0A1XK87 T1PEI7 T1PG43 T1PLK6 E0W0P8 A0A0A1WF26 A0A034V805 A0A034V5X0 A0A1I8NAM0 A0A1L8DMU0 A0A1Q3FTQ1 A0A1Q3FTS8 A0A1Q3FTL7 A0A1Q3FTM3 A0A1Q3FTL2 A0A146KTI9 A0A034V709 A0A034V3N2 A0A1W4UTM9 A0A2J7QEF9 A0A2J7QED8 A0A0Q5U557 A0A0J9RXH5 A0A1W4UTX3 Q86R99 A8JNT5 A0A1W4UG54 A0A0Q5UH57 A0A0J9RW24 A0A0Q5U8Y8 A0A1W4UTW8 A0A0R3P353 A0A0J9RW22 A0A0R3P811 A0A0R3P3C7 A0A1W4UGQ2 M9PCQ3 Q8MV37 A0A0P8XVK7 A0A0Q9WQJ7 A0A0Q9XBS6 B4L081 A0A0Q9XBJ6 A0A084WA83 A0A0P8XVL6 A0A0Q9WI93 A0A0P8XVM6 A0A1W4UGP7 A0A1W4UVB6 A0A0J9RWE5 A0A2S2QF17 B4HJ07 B3NDR1 Q8IQN4 Q8MT32 K7J9N7 A0A0M4ECR6 A0A0C9RGS4 A0A1B6JXK3 A0A146KR34 A0A1B6IZ70 A0A1B6GT14 A0A146M082
PDB
1S70
E-value=5.07939e-71,
Score=683
Ontologies
GO
GO:0019901
GO:0007165
GO:0019208
GO:0032440
GO:0016021
GO:0008544
GO:0035317
GO:0007391
GO:0045314
GO:0004857
GO:1904059
GO:0048675
GO:0045179
GO:0007560
GO:0030725
GO:0005737
GO:0048812
GO:0045177
GO:0043296
GO:0005829
GO:0005515
GO:0004930
GO:0007186
GO:0016020
GO:0015074
GO:0048013
GO:0005003
GO:0005887
GO:0007169
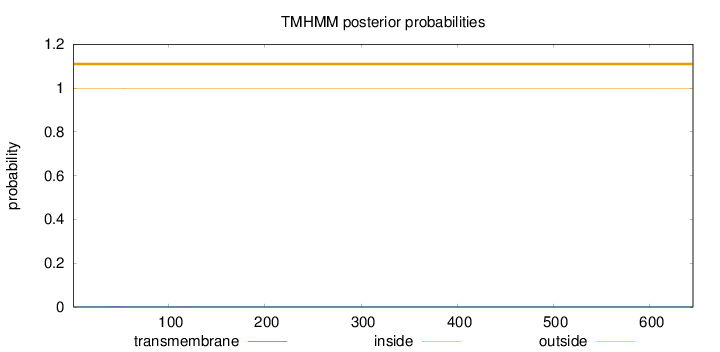
Topology
Length:
645
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02848
Exp number, first 60 AAs:
0.02354
Total prob of N-in:
0.00203
outside
1 - 645
Population Genetic Test Statistics
Pi
19.765462
Theta
20.020585
Tajima's D
-1.021385
CLR
0.094439
CSRT
0.135643217839108
Interpretation
Uncertain
