Gene
KWMTBOMO00127
Pre Gene Modal
BGIBMGA002138
Annotation
PREDICTED:_CUB_and_sushi_domain-containing_protein_2_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 3.369
Sequence
CDS
ATGCTCTACAACATAATATTTTGCTCTACCGCCGCAACCAAGTCTTCTCCAACAACGCTAGAATAGCGTCGCTGTGGTCATCCTGCAGTACCGCCAAACGCAAGAGTATCAATACCCAATGAAGAAAACATCGTTGCTGGAACCATAGCCGTCTATGAATGCGACGAAGGATATGAACTGTTCGGAGCTCGCCAGCGGGAGTGCACCTTAAGAGGAGACTGGACCCTAGAACCGCCTTTCTGTGGAACGAATGTGGCCTTCAGGAAGCCTGCCAATCAGTCTACAACGGTTCGAGGAGGTGCAGCAGCTAATGGCAACGATGGAGAGAAAACCACTGAGCATGACCGCAAGAGGTGCACGGAGACACAGCGAGAAGCGTCTCCGTGGTGGCAAGTTGATCTGCTCAGGCATTATGCTATCAAGGTCGTCAGGGTCACAACACGAGGCTGCTGTGGCCATCAACCTCTTCAAGAGTTGGAGATCAGAGTCGGAAACAGCAGTTCTGACCTCCAGCGGAACCCTCTATGTGCCTGGTTTCCTGGAACTATTGACGAAGGCGTTACGAAAACGTTCACGTGCGCTCGACCTCTGATCGGGCAGCACGTGTTTCTGCAACTGGTTGGTGTCGAGGGCTCCTTATCGCTTTGCGAAGTTGAAGTTTTTACAACAGAAGAATTTTCGAACGATCGATGCGCTCCGGCCGGCGCCCCCGCTGATATTGAATTGGCTGCGTTCTCCAGAAACTGTTATGAGTTCAACGTTGCCAGGGGGGGCTCGTTCGACGAAGCCAGGAAGCAATGTCAATCACATGGTGGCGATTTAATACACGGCTTTCAGGGTGCCACGTCGAGTTTCCTCACTCAAGAACTGGAGCGTCGTAAAGCACAGTTAAAAACACAGTTGGTCTGGATCGGAGCCCAAAAGGAGCCGGGCCTCACATCTCGAACCTGGCAATGGGTGGATGGCGAATTAGTTTCGAAGCCGAGCTGGGGTAAAGACCAGCCTAATAATTACAACGGGGAACAGAACTGCGTTGTCCTGGACGGCGGGAGAGGCTGGTTGTGGAACGACGTTGGCTGCAACCTCGATTACTTGCACTGGATCTGCCAGTACCTGCCTCCCACGTGCGGCAGTCCGGATAAACTACTGAATACTACCATACTCGGAAACGATTATGATGTTGGATCAAGAATTGTCTACAAGTGCCCCGAAGGTCATATGTTGCTCGGCGACAAAAGCAGGCACTGCAAGAAAGACGGGTTCTGGTCTGGAGCACCTCCCAGCTGTAAATATGTAAATTGTGGGGGTCTAACGCCGATCGAAGACGGAGATGTGGCTCTTATTGACAACAGAACAACCCACGGGGCTAGAGCTCTTTACTCCTGCCGCGAAAACTTCACCTTAGTCGGAAACCCTGAAAGAACCTGCGGTGATGAAGGTATTTGGGATGGAGAGGCTCCGAGATGCCTGTTCGACTGGTGTCCCGATCCTCCCGCCGTCTCCAACGCCTCAGTGACTGTCAATGGACACAAAGCGGGTTCTTTGGCTTCGTATACGTGTGAAAATGGCCATATTCTTTCCGGGTCAACGAGCGTCACTTGCACCCTCGGTGGTGCTTGGTCAGGAGTACCTCCGTCCTGTAAATACGTCAACTGTGGAACCCCGACAATTCACAAGGGCTCTTTTGTACTCATCAACGGAACCACCACATATGGCTCCATTGCCCAGTTCACATGCGAGCCCGACTACTGGTTGAACGGAGCTGAAGAGCTGGTTTGCAATAGGGACGGCAAGTGGTCGTATGACATCCCATCTTGTGAATTGACATCGTGTCCAGAGCCAGAAGTACCTTCCGGCGGTTACATGGAGGCTTTCGATTACTACGTGCACTCAACAATAAACTTCCACTGCGAAGCGGGACATAAGCTGATAGGGGAACCCAATCTCGTTTGCCAGCAAGACGGAGAATGGTCAGGGGATTCGCCGACATGTGAATATATCGATTGCGGGAAATTGCCAACATTGCCGTACGGCACAATGGAACTGTTAAACGGAACGACCCACCTGGGAAGCGTCATTCAGTACTCATGCACGACCAACTACAGACTTGTTGGTCCAGTAAGGAGGACCTGCACCGAAGAATTTCAATGGAGCGATTCTTCGCCCAGATGTGAAGAAATCCGATGTTCCGATCCGATAGTGGCCGAGAAGAGCATAGTATCCGTGACCGGTAACGATAGGAGCCATGGGCACACGCTGATCAGGACGTCTGACAACAGAGACGTCGGAAACACTTACCGTATTGGAGCGCTTGTGAAGTACAGATGTGAGAAAGGATACAAAGTGATCGGAGAGAGCTTATCGACTTGTGAAGATAATGGACAGTGGAGTGGAACAACGCCAACCTGCCAATACGTCGACTGCGGCAATCCAGGACGAATCGAGAATGGCAAAGTGACTCTTCCATCGAACGCCACATTCTATGGGGCCACCGCTTTCTACCAATGTGATGAAAACTGGGAGCTGGAGGGCGTGATCAGGAGAATGTGCCAAGAGAATGGGACTTGGAGTTTGGAGCCTCCCATTTGCAAAGAGATCACCTGCCCGGACCCATCGGCCCAAATCAAGGCCGGATCTGGTCTGTTAGTTGTTACGTCGACCCTCAGCATTGGGGGAGAGGCCCATTACCAGTGCGAGAGAGGTTACAACCTCAAAGGTCACCAGACAAGAACCTGCATATCGAAAGGACAGTGGAATGGAACTGTGCCTGTTTGCATACCAATCGACTGTAGGTCACCGGGAAATATAGAGAACGGTAAAATCGTCATCTCTAACGGCTCCACGCTGTTCGGAAGCTCCGTGGAATATTACTGTCTATCCCAATACCAGCGAGTTGGACCTTTCTTGAGAAAGTGTCTGGATGACGGTCGCTGGTCGGGTGAAGAGCCAAGCTGTGAACTAGCGACCAACGAGGCTGTGGACAGTGGAACTCTGCCCCTAAGTGTTGGGGTCGGGTGTGGAGTGGTCCTGTTCCTACTGATGCTTCTAGGAATCGTTTATCTGAGGCTACGGAAAGCAACGCCTGTTAAGAACACGGAAAACATTGAGGGCGCCGAGCGAAAGGAAGATCAGAGCGCGGCGGTGATGAGCTACGCCACGTTGCACGACGCCAGCGGACGCCACATCTACGACCACGTTTCTGACAACGTGTACGACTCGCCTTACAGCGAACAGATAACTGACAACGTCGCGTACGGCAGACGAGGCGACACCGACTCCGCCTACGAGCCAGAGCCGACCGGACCCAATGCTGTCATCACCATCAACGGAGTCGCGGTGCGTTGA
Protein
MLYNIIFCSTAATKSSPTTLEXRRCGHPAVPPNARVSIPNEENIVAGTIAVYECDEGYELFGARQRECTLRGDWTLEPPFCGTNVAFRKPANQSTTVRGGAAANGNDGEKTTEHDRKRCTETQREASPWWQVDLLRHYAIKVVRVTTRGCCGHQPLQELEIRVGNSSSDLQRNPLCAWFPGTIDEGVTKTFTCARPLIGQHVFLQLVGVEGSLSLCEVEVFTTEEFSNDRCAPAGAPADIELAAFSRNCYEFNVARGGSFDEARKQCQSHGGDLIHGFQGATSSFLTQELERRKAQLKTQLVWIGAQKEPGLTSRTWQWVDGELVSKPSWGKDQPNNYNGEQNCVVLDGGRGWLWNDVGCNLDYLHWICQYLPPTCGSPDKLLNTTILGNDYDVGSRIVYKCPEGHMLLGDKSRHCKKDGFWSGAPPSCKYVNCGGLTPIEDGDVALIDNRTTHGARALYSCRENFTLVGNPERTCGDEGIWDGEAPRCLFDWCPDPPAVSNASVTVNGHKAGSLASYTCENGHILSGSTSVTCTLGGAWSGVPPSCKYVNCGTPTIHKGSFVLINGTTTYGSIAQFTCEPDYWLNGAEELVCNRDGKWSYDIPSCELTSCPEPEVPSGGYMEAFDYYVHSTINFHCEAGHKLIGEPNLVCQQDGEWSGDSPTCEYIDCGKLPTLPYGTMELLNGTTHLGSVIQYSCTTNYRLVGPVRRTCTEEFQWSDSSPRCEEIRCSDPIVAEKSIVSVTGNDRSHGHTLIRTSDNRDVGNTYRIGALVKYRCEKGYKVIGESLSTCEDNGQWSGTTPTCQYVDCGNPGRIENGKVTLPSNATFYGATAFYQCDENWELEGVIRRMCQENGTWSLEPPICKEITCPDPSAQIKAGSGLLVVTSTLSIGGEAHYQCERGYNLKGHQTRTCISKGQWNGTVPVCIPIDCRSPGNIENGKIVISNGSTLFGSSVEYYCLSQYQRVGPFLRKCLDDGRWSGEEPSCELATNEAVDSGTLPLSVGVGCGVVLFLLMLLGIVYLRLRKATPVKNTENIEGAERKEDQSAAVMSYATLHDASGRHIYDHVSDNVYDSPYSEQITDNVAYGRRGDTDSAYEPEPTGPNAVITINGVAVR
Summary
Uniprot
H9IY05
A0A194QUA4
A0A194Q4W1
A0A2A4K0P2
A0A212F9V1
A0A219YXI7
+ More
A0A2H1WY99 A0A1V1FYC5 D6WIC1 A0A2A3EGS2 E2BVC7 E2ASL9 A0A195BZZ9 A0A0L7QSA9 A0A088AU37 A0A154NYR3 A0A1S4MZJ1 A0A0N0BJ10 A0A195F0U9 A0A195EGB6 A0A151XBK3 A0A146M788 A0A158NL28 A0A195BHB5 A0A0C9QVS0 A0A1B6G943 F4WFY7 E0VZV6 A0A0C9RHU4 A0A0K8SU93 K7IN02 A0A232FA27 T1HQV6 A0A067R1P5 A0A1B0CZ92 A0A2J7RKX7 A0A2H8TPR4 A0A2S2NGM5 A0A0L0BT81 J9JRW4 T1P8S0 A0A0K8VIV7 A0A1I8PKJ5 A0A0K8VMI0 A0A034V0Z7 A0A1I8MCU6 W8AQB1 A0A182UU82 A0A182IDJ5 A0A182WRB3 A0A1S4GAG1 Q7QI37 A0A182RKB4 A0A084WPQ7 A0A182KFP5 A0A182NVF0 A0A1B0FNU2 A0A182VT36 A0A1A9W649 A0A1A9VUL7 A0A182FK45 A0A182QIU9 A0A182MSV8 A0A182PL41 A0A182YQ35 A0A2M4CHZ4 B3NXM9 A0A182J2C0 A0A1J1IPH3 B4PYN4 B4L1H8 B4ILA4 Q9VYR4 B5DNH9 A0A3B0KQ61 A0A0M4FB02 A0A1W4VUG9 A0A1W4VTQ8 B4M234 A0A1W4VH46 A0A1B0A0W9 N6UB14 B3N0R0 B4ND16 A0A026WXD4 Q16XC0 B4JIR0 A0A224XJS3 W5JFD4 A0A1A9YF17 A0A0A9Y606 A0A1B0AMS5 A0A3L8D808 B4H0Z0 A0A1B6DUN5 A0A336K2F8 A0A1Y1MWR4 A0A2P8YK25 A0A147BFW2
A0A2H1WY99 A0A1V1FYC5 D6WIC1 A0A2A3EGS2 E2BVC7 E2ASL9 A0A195BZZ9 A0A0L7QSA9 A0A088AU37 A0A154NYR3 A0A1S4MZJ1 A0A0N0BJ10 A0A195F0U9 A0A195EGB6 A0A151XBK3 A0A146M788 A0A158NL28 A0A195BHB5 A0A0C9QVS0 A0A1B6G943 F4WFY7 E0VZV6 A0A0C9RHU4 A0A0K8SU93 K7IN02 A0A232FA27 T1HQV6 A0A067R1P5 A0A1B0CZ92 A0A2J7RKX7 A0A2H8TPR4 A0A2S2NGM5 A0A0L0BT81 J9JRW4 T1P8S0 A0A0K8VIV7 A0A1I8PKJ5 A0A0K8VMI0 A0A034V0Z7 A0A1I8MCU6 W8AQB1 A0A182UU82 A0A182IDJ5 A0A182WRB3 A0A1S4GAG1 Q7QI37 A0A182RKB4 A0A084WPQ7 A0A182KFP5 A0A182NVF0 A0A1B0FNU2 A0A182VT36 A0A1A9W649 A0A1A9VUL7 A0A182FK45 A0A182QIU9 A0A182MSV8 A0A182PL41 A0A182YQ35 A0A2M4CHZ4 B3NXM9 A0A182J2C0 A0A1J1IPH3 B4PYN4 B4L1H8 B4ILA4 Q9VYR4 B5DNH9 A0A3B0KQ61 A0A0M4FB02 A0A1W4VUG9 A0A1W4VTQ8 B4M234 A0A1W4VH46 A0A1B0A0W9 N6UB14 B3N0R0 B4ND16 A0A026WXD4 Q16XC0 B4JIR0 A0A224XJS3 W5JFD4 A0A1A9YF17 A0A0A9Y606 A0A1B0AMS5 A0A3L8D808 B4H0Z0 A0A1B6DUN5 A0A336K2F8 A0A1Y1MWR4 A0A2P8YK25 A0A147BFW2
Pubmed
19121390
26354079
22118469
28410430
18362917
19820115
+ More
20798317 20566863 26823975 21347285 21719571 20075255 28648823 24845553 26108605 25348373 25315136 24495485 12364791 24438588 25244985 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23537049 24508170 17510324 20920257 23761445 25401762 30249741 28004739 29403074
20798317 20566863 26823975 21347285 21719571 20075255 28648823 24845553 26108605 25348373 25315136 24495485 12364791 24438588 25244985 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23537049 24508170 17510324 20920257 23761445 25401762 30249741 28004739 29403074
EMBL
BABH01015930
BABH01015931
KQ461108
KPJ09108.1
KQ459463
KPJ00573.1
+ More
NWSH01000275 PCG77845.1 AGBW02009564 OWR50489.1 KX550143 AQY54460.1 ODYU01011554 SOQ57364.1 FX985256 BAX07269.1 KQ971321 EFA01446.2 KZ288255 PBC30674.1 GL450824 EFN80419.1 GL442330 EFN63579.1 KQ978501 KYM93483.1 KQ414758 KOC61518.1 KQ434783 KZC04763.1 AAZO01006540 AAZO01006541 AAZO01006542 AAZO01006543 AAZO01006544 KQ435724 KOX78438.1 KQ981866 KYN34103.1 KQ978957 KYN26934.1 KQ982320 KYQ57755.1 GDHC01003330 JAQ15299.1 ADTU01019226 ADTU01019227 ADTU01019228 KQ976488 KYM83544.1 GBYB01007834 JAG77601.1 GECZ01010797 JAS58972.1 GL888128 EGI66754.1 DS235854 EEB18912.1 GBYB01007835 JAG77602.1 GBRD01008948 JAG56873.1 NNAY01000629 OXU27309.1 ACPB03023230 KK852985 KDR12825.1 AJVK01002143 AJVK01002144 AJVK01002145 AJVK01002146 NEVH01002704 PNF41493.1 GFXV01004194 MBW15999.1 GGMR01003463 MBY16082.1 JRES01001383 KNC23231.1 ABLF02032708 KA645072 AFP59701.1 GDHF01013488 JAI38826.1 GDHF01012579 JAI39735.1 GAKP01022002 GAKP01022001 JAC36951.1 GAMC01018303 GAMC01018302 JAB88252.1 APCN01004705 APCN01004706 APCN01004707 AAAB01008811 EAA04956.3 ATLV01025119 KE525369 KFB52201.1 CCAG010009643 AXCN02002011 AXCM01003211 GGFL01000788 MBW64966.1 CH954180 EDV47330.2 CVRI01000055 CRL01626.1 CM000162 EDX00970.1 CH933810 EDW07614.2 CH480868 EDW53750.1 AE014298 BT003214 AAF48125.2 AAO24969.1 AHN59616.1 CH379064 EDY72987.2 OUUW01000015 SPP88759.1 CP012528 ALC49695.1 CH940651 EDW65738.2 APGK01041924 APGK01041925 KB740998 ENN75827.1 CH902644 EDV34789.2 CH964239 EDW82725.2 KK107078 EZA60406.1 CH477543 EAT39254.2 CH916370 EDW00507.1 GFTR01008155 JAW08271.1 ADMH02001566 ETN62048.1 GBHO01016543 JAG27061.1 JXJN01000557 QOIP01000011 RLU16414.1 CH479201 EDW29906.1 GEDC01007916 JAS29382.1 UFQS01000005 UFQT01000005 SSW96933.1 SSX17320.1 GEZM01021615 JAV88905.1 PYGN01000541 PSN44564.1 GEGO01005753 JAR89651.1
NWSH01000275 PCG77845.1 AGBW02009564 OWR50489.1 KX550143 AQY54460.1 ODYU01011554 SOQ57364.1 FX985256 BAX07269.1 KQ971321 EFA01446.2 KZ288255 PBC30674.1 GL450824 EFN80419.1 GL442330 EFN63579.1 KQ978501 KYM93483.1 KQ414758 KOC61518.1 KQ434783 KZC04763.1 AAZO01006540 AAZO01006541 AAZO01006542 AAZO01006543 AAZO01006544 KQ435724 KOX78438.1 KQ981866 KYN34103.1 KQ978957 KYN26934.1 KQ982320 KYQ57755.1 GDHC01003330 JAQ15299.1 ADTU01019226 ADTU01019227 ADTU01019228 KQ976488 KYM83544.1 GBYB01007834 JAG77601.1 GECZ01010797 JAS58972.1 GL888128 EGI66754.1 DS235854 EEB18912.1 GBYB01007835 JAG77602.1 GBRD01008948 JAG56873.1 NNAY01000629 OXU27309.1 ACPB03023230 KK852985 KDR12825.1 AJVK01002143 AJVK01002144 AJVK01002145 AJVK01002146 NEVH01002704 PNF41493.1 GFXV01004194 MBW15999.1 GGMR01003463 MBY16082.1 JRES01001383 KNC23231.1 ABLF02032708 KA645072 AFP59701.1 GDHF01013488 JAI38826.1 GDHF01012579 JAI39735.1 GAKP01022002 GAKP01022001 JAC36951.1 GAMC01018303 GAMC01018302 JAB88252.1 APCN01004705 APCN01004706 APCN01004707 AAAB01008811 EAA04956.3 ATLV01025119 KE525369 KFB52201.1 CCAG010009643 AXCN02002011 AXCM01003211 GGFL01000788 MBW64966.1 CH954180 EDV47330.2 CVRI01000055 CRL01626.1 CM000162 EDX00970.1 CH933810 EDW07614.2 CH480868 EDW53750.1 AE014298 BT003214 AAF48125.2 AAO24969.1 AHN59616.1 CH379064 EDY72987.2 OUUW01000015 SPP88759.1 CP012528 ALC49695.1 CH940651 EDW65738.2 APGK01041924 APGK01041925 KB740998 ENN75827.1 CH902644 EDV34789.2 CH964239 EDW82725.2 KK107078 EZA60406.1 CH477543 EAT39254.2 CH916370 EDW00507.1 GFTR01008155 JAW08271.1 ADMH02001566 ETN62048.1 GBHO01016543 JAG27061.1 JXJN01000557 QOIP01000011 RLU16414.1 CH479201 EDW29906.1 GEDC01007916 JAS29382.1 UFQS01000005 UFQT01000005 SSW96933.1 SSX17320.1 GEZM01021615 JAV88905.1 PYGN01000541 PSN44564.1 GEGO01005753 JAR89651.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000007266
+ More
UP000242457 UP000008237 UP000000311 UP000078542 UP000053825 UP000005203 UP000076502 UP000053105 UP000078541 UP000078492 UP000075809 UP000005205 UP000078540 UP000007755 UP000009046 UP000002358 UP000215335 UP000015103 UP000027135 UP000092462 UP000235965 UP000037069 UP000007819 UP000095300 UP000095301 UP000075903 UP000075840 UP000076407 UP000007062 UP000075900 UP000030765 UP000075881 UP000075884 UP000092444 UP000075920 UP000091820 UP000078200 UP000069272 UP000075886 UP000075883 UP000075885 UP000076408 UP000008711 UP000075880 UP000183832 UP000002282 UP000009192 UP000001292 UP000000803 UP000001819 UP000268350 UP000092553 UP000192221 UP000008792 UP000092445 UP000019118 UP000007801 UP000007798 UP000053097 UP000008820 UP000001070 UP000000673 UP000092443 UP000092460 UP000279307 UP000008744 UP000245037
UP000242457 UP000008237 UP000000311 UP000078542 UP000053825 UP000005203 UP000076502 UP000053105 UP000078541 UP000078492 UP000075809 UP000005205 UP000078540 UP000007755 UP000009046 UP000002358 UP000215335 UP000015103 UP000027135 UP000092462 UP000235965 UP000037069 UP000007819 UP000095300 UP000095301 UP000075903 UP000075840 UP000076407 UP000007062 UP000075900 UP000030765 UP000075881 UP000075884 UP000092444 UP000075920 UP000091820 UP000078200 UP000069272 UP000075886 UP000075883 UP000075885 UP000076408 UP000008711 UP000075880 UP000183832 UP000002282 UP000009192 UP000001292 UP000000803 UP000001819 UP000268350 UP000092553 UP000192221 UP000008792 UP000092445 UP000019118 UP000007801 UP000007798 UP000053097 UP000008820 UP000001070 UP000000673 UP000092443 UP000092460 UP000279307 UP000008744 UP000245037
PRIDE
Interpro
IPR018378
C-type_lectin_CS
+ More
IPR008979 Galactose-bd-like_sf
IPR016187 CTDL_fold
IPR000436 Sushi_SCR_CCP_dom
IPR016186 C-type_lectin-like/link_sf
IPR035976 Sushi/SCR/CCP_sf
IPR001304 C-type_lectin-like
IPR006585 FTP1
IPR000421 FA58C
IPR001879 GPCR_2_extracellular_dom
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR018247 EF_Hand_1_Ca_BS
IPR007110 Ig-like_dom
IPR008979 Galactose-bd-like_sf
IPR016187 CTDL_fold
IPR000436 Sushi_SCR_CCP_dom
IPR016186 C-type_lectin-like/link_sf
IPR035976 Sushi/SCR/CCP_sf
IPR001304 C-type_lectin-like
IPR006585 FTP1
IPR000421 FA58C
IPR001879 GPCR_2_extracellular_dom
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR018247 EF_Hand_1_Ca_BS
IPR007110 Ig-like_dom
Gene 3D
ProteinModelPortal
H9IY05
A0A194QUA4
A0A194Q4W1
A0A2A4K0P2
A0A212F9V1
A0A219YXI7
+ More
A0A2H1WY99 A0A1V1FYC5 D6WIC1 A0A2A3EGS2 E2BVC7 E2ASL9 A0A195BZZ9 A0A0L7QSA9 A0A088AU37 A0A154NYR3 A0A1S4MZJ1 A0A0N0BJ10 A0A195F0U9 A0A195EGB6 A0A151XBK3 A0A146M788 A0A158NL28 A0A195BHB5 A0A0C9QVS0 A0A1B6G943 F4WFY7 E0VZV6 A0A0C9RHU4 A0A0K8SU93 K7IN02 A0A232FA27 T1HQV6 A0A067R1P5 A0A1B0CZ92 A0A2J7RKX7 A0A2H8TPR4 A0A2S2NGM5 A0A0L0BT81 J9JRW4 T1P8S0 A0A0K8VIV7 A0A1I8PKJ5 A0A0K8VMI0 A0A034V0Z7 A0A1I8MCU6 W8AQB1 A0A182UU82 A0A182IDJ5 A0A182WRB3 A0A1S4GAG1 Q7QI37 A0A182RKB4 A0A084WPQ7 A0A182KFP5 A0A182NVF0 A0A1B0FNU2 A0A182VT36 A0A1A9W649 A0A1A9VUL7 A0A182FK45 A0A182QIU9 A0A182MSV8 A0A182PL41 A0A182YQ35 A0A2M4CHZ4 B3NXM9 A0A182J2C0 A0A1J1IPH3 B4PYN4 B4L1H8 B4ILA4 Q9VYR4 B5DNH9 A0A3B0KQ61 A0A0M4FB02 A0A1W4VUG9 A0A1W4VTQ8 B4M234 A0A1W4VH46 A0A1B0A0W9 N6UB14 B3N0R0 B4ND16 A0A026WXD4 Q16XC0 B4JIR0 A0A224XJS3 W5JFD4 A0A1A9YF17 A0A0A9Y606 A0A1B0AMS5 A0A3L8D808 B4H0Z0 A0A1B6DUN5 A0A336K2F8 A0A1Y1MWR4 A0A2P8YK25 A0A147BFW2
A0A2H1WY99 A0A1V1FYC5 D6WIC1 A0A2A3EGS2 E2BVC7 E2ASL9 A0A195BZZ9 A0A0L7QSA9 A0A088AU37 A0A154NYR3 A0A1S4MZJ1 A0A0N0BJ10 A0A195F0U9 A0A195EGB6 A0A151XBK3 A0A146M788 A0A158NL28 A0A195BHB5 A0A0C9QVS0 A0A1B6G943 F4WFY7 E0VZV6 A0A0C9RHU4 A0A0K8SU93 K7IN02 A0A232FA27 T1HQV6 A0A067R1P5 A0A1B0CZ92 A0A2J7RKX7 A0A2H8TPR4 A0A2S2NGM5 A0A0L0BT81 J9JRW4 T1P8S0 A0A0K8VIV7 A0A1I8PKJ5 A0A0K8VMI0 A0A034V0Z7 A0A1I8MCU6 W8AQB1 A0A182UU82 A0A182IDJ5 A0A182WRB3 A0A1S4GAG1 Q7QI37 A0A182RKB4 A0A084WPQ7 A0A182KFP5 A0A182NVF0 A0A1B0FNU2 A0A182VT36 A0A1A9W649 A0A1A9VUL7 A0A182FK45 A0A182QIU9 A0A182MSV8 A0A182PL41 A0A182YQ35 A0A2M4CHZ4 B3NXM9 A0A182J2C0 A0A1J1IPH3 B4PYN4 B4L1H8 B4ILA4 Q9VYR4 B5DNH9 A0A3B0KQ61 A0A0M4FB02 A0A1W4VUG9 A0A1W4VTQ8 B4M234 A0A1W4VH46 A0A1B0A0W9 N6UB14 B3N0R0 B4ND16 A0A026WXD4 Q16XC0 B4JIR0 A0A224XJS3 W5JFD4 A0A1A9YF17 A0A0A9Y606 A0A1B0AMS5 A0A3L8D808 B4H0Z0 A0A1B6DUN5 A0A336K2F8 A0A1Y1MWR4 A0A2P8YK25 A0A147BFW2
PDB
2Q7Z
E-value=3.41856e-53,
Score=531
Ontologies
GO
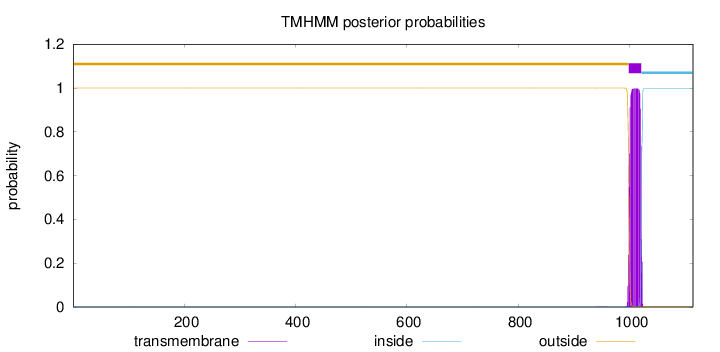
Topology
Length:
1114
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.56444
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00003
outside
1 - 998
TMhelix
999 - 1021
inside
1022 - 1114
Population Genetic Test Statistics
Pi
26.06811
Theta
23.646185
Tajima's D
-1.305989
CLR
0.019141
CSRT
0.0875456227188641
Interpretation
Uncertain
