Gene
KWMTBOMO00125
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.608
Sequence
CDS
ATGGTCTACCTGGTCCCTCGTTTGTTGGGGGTGGCCGGCGCGCTAAGCCGGCTTCTGCCCAACGTCGGGGGGCCTGACCAGGTGACGCGCCGTCTCTATACGGGGGTGGTGCGATCAATGGCCCTATACGGGGCGCCCGTGTGGGGCCAGTCCCTGGCCGTGGGGGTGGCGAAGCTGCTGCAACGGCCGCAACGCACCATCGCGGTCAGGGTCATCCGTGGTTATCGCACCATCTCCTTTGAGGCGGCGTGTGTACTGGCTGGGACGCCGCCTTGGGTTCTGGAAGAGGAGGCGCTCGCCGCTGACTATAGGTGGCGGGCTGAACTTCGTACCTGGGGCGTGGCGCGTCCCAGCCCCAGTGTGGTCAGAGCACGGAGGGCCCAATCTCGGCGGTCCGTGCTGGAGTCATGGTCCAGACGGCTGGCTGATCCTTCGGCTGGTCGTAGGACCGTCGAGGCGATTCGCCCGGTCCTTGTGGATTGGGTGAATCGTGACAGAGGACGCCTCACTTTCCGGCTTACGCAGGTGCTCACTGGGCATGGTTGTTTCGGTGAGTTCCTGCACCGGATCACAGCCGAGCCGACGGCAGAGTGCCACCATTGTGGTTGCGACTTGGACACGGCAGAGCATACGCTCGTCGCCTGCCCCGCATGGGAGGGGTGGCGCCGTGTCCTCGTCGCAAAAATAGGAACCGACTTGTCGTTGCCGAGTGTTGTGGCATCGATGCTCGGCGACGACGAGTCGTGGAAGGCGATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGGGCGCGTGAGAGACGCACAATCCCGCCGCCGTCGAGCGGGGGCCAGGGAGGCGGATCTCGCCCAAGCCCTGGCCCTCTAA
Protein
MVYLVPRLLGVAGALSRLLPNVGGPDQVTRRLYTGVVRSMALYGAPVWGQSLAVGVAKLLQRPQRTIAVRVIRGYRTISFEAACVLAGTPPWVLEEEALAADYRWRAELRTWGVARPSPSVVRARRAQSRRSVLESWSRRLADPSAGRRTVEAIRPVLVDWVNRDRGRLTFRLTQVLTGHGCFGEFLHRITAEPTAECHHCGCDLDTAEHTLVACPAWEGWRRVLVAKIGTDLSLPSVVASMLGDDESWKAMLDFCECTISQKEAAGRVRDAQSRRRRAGAREADLAQALAL
Summary
Uniprot
Q5KTM7
Q868Q4
A0A2W1B333
A0A2W1BDU5
A0A2W1BSQ5
Q8MY27
+ More
Q8MY31 Q8MY33 Q8MY35 A0A2W1C3J5 A0A0J7KCI1 O01419 A0A0J7N1D9 A0A0J7N7H9 A0A0J7KCQ3 A0A0J7MY07 A0A0J7MZM4 A0A0J7KS67 A0A0J7KJB9 E2BVR6 A0A0J7KM11 A0A194R5D3 A0A0J7KIY3 A0A0J7KJH4 A0A0J7KIY5 A0A2H1WNZ5 A0A0J7KLT9 A0A0J7JZF7 A0A0J7JZ13 A0A0L7R7C3 A0A0J7KJG6 A0A0J7N7S8 A0A0J7JXN3 A0A0J7NEG9 A0A0J7KQK3 A0A3L8DIL1 A0A3L8DDV9 A0A0J7MX22 A0A3L8D2G3 A0A0J7KFG9 A0A0J7KIW1 A0A2S2NPF9 A0A0J7KIM6 A0A2S2PJ67 A0A0J7KPR0 A0A0J7K3F1 A0A0J7KYD2 J9KRD0 A0A0J7K840 A0A0J7KWU5 A0A0J7JYP2 A0A2S2P6S8 A0A0J7KPZ3 J9KJG8 A0A0J7KRB8 E2A7V5 A0A0J7NCZ4 A0A0J7NMN5 A0A0J7NAV8 A0A0J7KL06 A0A2H8TR36 A0A0J7KC11 A0A2S2NME4 A0A2S2PYG9 J9KLZ3 A0A2S2PSC1 A0A2S2NJ82 A0A3Q0JIB3 X1WJY4 A0A0J7K6Z4 A0A0J7KRU6 A0A0Q9WRU0 A0A0J7MY17 A0A2S2PEU2 J9LAC1 A0A2H8THF8 J9LDW8 A0A2S2PDK9 A0A0J7KIA8 A0A0J7KF40 J9LJ80 A0A2S2PEZ7 A0A0J7KLH8 A0A0J7KKI5 X1WPJ7 A0A0J7NFZ0 J9KNG8
Q8MY31 Q8MY33 Q8MY35 A0A2W1C3J5 A0A0J7KCI1 O01419 A0A0J7N1D9 A0A0J7N7H9 A0A0J7KCQ3 A0A0J7MY07 A0A0J7MZM4 A0A0J7KS67 A0A0J7KJB9 E2BVR6 A0A0J7KM11 A0A194R5D3 A0A0J7KIY3 A0A0J7KJH4 A0A0J7KIY5 A0A2H1WNZ5 A0A0J7KLT9 A0A0J7JZF7 A0A0J7JZ13 A0A0L7R7C3 A0A0J7KJG6 A0A0J7N7S8 A0A0J7JXN3 A0A0J7NEG9 A0A0J7KQK3 A0A3L8DIL1 A0A3L8DDV9 A0A0J7MX22 A0A3L8D2G3 A0A0J7KFG9 A0A0J7KIW1 A0A2S2NPF9 A0A0J7KIM6 A0A2S2PJ67 A0A0J7KPR0 A0A0J7K3F1 A0A0J7KYD2 J9KRD0 A0A0J7K840 A0A0J7KWU5 A0A0J7JYP2 A0A2S2P6S8 A0A0J7KPZ3 J9KJG8 A0A0J7KRB8 E2A7V5 A0A0J7NCZ4 A0A0J7NMN5 A0A0J7NAV8 A0A0J7KL06 A0A2H8TR36 A0A0J7KC11 A0A2S2NME4 A0A2S2PYG9 J9KLZ3 A0A2S2PSC1 A0A2S2NJ82 A0A3Q0JIB3 X1WJY4 A0A0J7K6Z4 A0A0J7KRU6 A0A0Q9WRU0 A0A0J7MY17 A0A2S2PEU2 J9LAC1 A0A2H8THF8 J9LDW8 A0A2S2PDK9 A0A0J7KIA8 A0A0J7KF40 J9LJ80 A0A2S2PEZ7 A0A0J7KLH8 A0A0J7KKI5 X1WPJ7 A0A0J7NFZ0 J9KNG8
EMBL
AB126050
BAD86650.1
AB090825
BAC57926.1
KZ150386
PZC71062.1
+ More
KZ150308 PZC71477.1 KZ149950 PZC76665.1 AB078935 BAC06462.1 AB078931 BAC06456.1 AB078930 BAC06454.1 AB078929 BAC06452.1 KZ149896 PZC78733.1 LBMM01009501 KMQ88083.1 D85594 BAA19776.1 LBMM01012027 KMQ86470.1 LBMM01008861 KMQ88580.1 LBMM01009494 KMQ88097.1 LBMM01014132 KMQ85305.1 LBMM01012957 LBMM01012956 KMQ85900.1 KMQ85901.1 LBMM01003679 KMQ93242.1 LBMM01006538 KMQ90523.1 GL450953 EFN80213.1 LBMM01005707 KMQ91279.1 KQ460685 KPJ12887.1 LBMM01006909 KMQ90209.1 LBMM01006657 KMQ90417.1 LBMM01006954 KMQ90166.1 ODYU01009945 SOQ54696.1 LBMM01005733 KMQ91252.1 LBMM01019838 KMQ83439.1 LBMM01019839 KMQ83438.1 KQ414641 KOC66782.1 LBMM01006671 KMQ90407.1 LBMM01008682 KMQ88705.1 LBMM01024024 KMQ82631.1 LBMM01006110 KMQ90925.1 LBMM01004272 KMQ92618.1 QOIP01000007 RLU20280.1 QOIP01000010 RLU18098.1 LBMM01014909 KMQ85000.1 QOIP01000031 RLU14697.1 LBMM01008349 KMQ88951.1 LBMM01006725 KMQ90373.1 GGMR01006442 MBY19061.1 LBMM01007036 KMQ90099.1 GGMR01016864 MBY29483.1 LBMM01004634 KMQ92224.1 LBMM01015025 KMQ84953.1 LBMM01002052 KMQ95336.1 ABLF02026677 ABLF02032994 ABLF02059021 ABLF02064578 LBMM01011877 KMQ86578.1 LBMM01002480 KMQ94776.1 LBMM01020849 KMQ83202.1 GGMR01012534 MBY25153.1 LBMM01004505 KMQ92346.1 ABLF02041886 LBMM01003988 KMQ92922.1 GL437417 EFN70484.1 LBMM01006640 KMQ90440.1 LBMM01003245 KMQ93755.1 LBMM01007425 KMQ89740.1 LBMM01006063 KMQ90967.1 GFXV01003903 MBW15708.1 LBMM01009738 KMQ87908.1 GGMR01005740 MBY18359.1 GGMS01001211 MBY70414.1 ABLF02023401 ABLF02029423 ABLF02060278 GGMR01019609 MBY32228.1 GGMR01004600 MBY17219.1 ABLF02011238 LBMM01012595 KMQ86127.1 LBMM01003941 KMQ92969.1 CH963857 KRF98317.1 LBMM01014122 KMQ85315.1 GGMR01015354 MBY27973.1 ABLF02017307 GFXV01001748 MBW13553.1 ABLF02016435 ABLF02016436 ABLF02064751 GGMR01014918 MBY27537.1 LBMM01007162 KMQ89979.1 LBMM01008422 KMQ88894.1 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 GGMR01015159 MBY27778.1 LBMM01005948 KMQ91066.1 LBMM01006287 KMQ90731.1 ABLF02013380 ABLF02013382 LBMM01005474 KMQ91485.1 ABLF02005203 ABLF02041316
KZ150308 PZC71477.1 KZ149950 PZC76665.1 AB078935 BAC06462.1 AB078931 BAC06456.1 AB078930 BAC06454.1 AB078929 BAC06452.1 KZ149896 PZC78733.1 LBMM01009501 KMQ88083.1 D85594 BAA19776.1 LBMM01012027 KMQ86470.1 LBMM01008861 KMQ88580.1 LBMM01009494 KMQ88097.1 LBMM01014132 KMQ85305.1 LBMM01012957 LBMM01012956 KMQ85900.1 KMQ85901.1 LBMM01003679 KMQ93242.1 LBMM01006538 KMQ90523.1 GL450953 EFN80213.1 LBMM01005707 KMQ91279.1 KQ460685 KPJ12887.1 LBMM01006909 KMQ90209.1 LBMM01006657 KMQ90417.1 LBMM01006954 KMQ90166.1 ODYU01009945 SOQ54696.1 LBMM01005733 KMQ91252.1 LBMM01019838 KMQ83439.1 LBMM01019839 KMQ83438.1 KQ414641 KOC66782.1 LBMM01006671 KMQ90407.1 LBMM01008682 KMQ88705.1 LBMM01024024 KMQ82631.1 LBMM01006110 KMQ90925.1 LBMM01004272 KMQ92618.1 QOIP01000007 RLU20280.1 QOIP01000010 RLU18098.1 LBMM01014909 KMQ85000.1 QOIP01000031 RLU14697.1 LBMM01008349 KMQ88951.1 LBMM01006725 KMQ90373.1 GGMR01006442 MBY19061.1 LBMM01007036 KMQ90099.1 GGMR01016864 MBY29483.1 LBMM01004634 KMQ92224.1 LBMM01015025 KMQ84953.1 LBMM01002052 KMQ95336.1 ABLF02026677 ABLF02032994 ABLF02059021 ABLF02064578 LBMM01011877 KMQ86578.1 LBMM01002480 KMQ94776.1 LBMM01020849 KMQ83202.1 GGMR01012534 MBY25153.1 LBMM01004505 KMQ92346.1 ABLF02041886 LBMM01003988 KMQ92922.1 GL437417 EFN70484.1 LBMM01006640 KMQ90440.1 LBMM01003245 KMQ93755.1 LBMM01007425 KMQ89740.1 LBMM01006063 KMQ90967.1 GFXV01003903 MBW15708.1 LBMM01009738 KMQ87908.1 GGMR01005740 MBY18359.1 GGMS01001211 MBY70414.1 ABLF02023401 ABLF02029423 ABLF02060278 GGMR01019609 MBY32228.1 GGMR01004600 MBY17219.1 ABLF02011238 LBMM01012595 KMQ86127.1 LBMM01003941 KMQ92969.1 CH963857 KRF98317.1 LBMM01014122 KMQ85315.1 GGMR01015354 MBY27973.1 ABLF02017307 GFXV01001748 MBW13553.1 ABLF02016435 ABLF02016436 ABLF02064751 GGMR01014918 MBY27537.1 LBMM01007162 KMQ89979.1 LBMM01008422 KMQ88894.1 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 GGMR01015159 MBY27778.1 LBMM01005948 KMQ91066.1 LBMM01006287 KMQ90731.1 ABLF02013380 ABLF02013382 LBMM01005474 KMQ91485.1 ABLF02005203 ABLF02041316
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
Q5KTM7
Q868Q4
A0A2W1B333
A0A2W1BDU5
A0A2W1BSQ5
Q8MY27
+ More
Q8MY31 Q8MY33 Q8MY35 A0A2W1C3J5 A0A0J7KCI1 O01419 A0A0J7N1D9 A0A0J7N7H9 A0A0J7KCQ3 A0A0J7MY07 A0A0J7MZM4 A0A0J7KS67 A0A0J7KJB9 E2BVR6 A0A0J7KM11 A0A194R5D3 A0A0J7KIY3 A0A0J7KJH4 A0A0J7KIY5 A0A2H1WNZ5 A0A0J7KLT9 A0A0J7JZF7 A0A0J7JZ13 A0A0L7R7C3 A0A0J7KJG6 A0A0J7N7S8 A0A0J7JXN3 A0A0J7NEG9 A0A0J7KQK3 A0A3L8DIL1 A0A3L8DDV9 A0A0J7MX22 A0A3L8D2G3 A0A0J7KFG9 A0A0J7KIW1 A0A2S2NPF9 A0A0J7KIM6 A0A2S2PJ67 A0A0J7KPR0 A0A0J7K3F1 A0A0J7KYD2 J9KRD0 A0A0J7K840 A0A0J7KWU5 A0A0J7JYP2 A0A2S2P6S8 A0A0J7KPZ3 J9KJG8 A0A0J7KRB8 E2A7V5 A0A0J7NCZ4 A0A0J7NMN5 A0A0J7NAV8 A0A0J7KL06 A0A2H8TR36 A0A0J7KC11 A0A2S2NME4 A0A2S2PYG9 J9KLZ3 A0A2S2PSC1 A0A2S2NJ82 A0A3Q0JIB3 X1WJY4 A0A0J7K6Z4 A0A0J7KRU6 A0A0Q9WRU0 A0A0J7MY17 A0A2S2PEU2 J9LAC1 A0A2H8THF8 J9LDW8 A0A2S2PDK9 A0A0J7KIA8 A0A0J7KF40 J9LJ80 A0A2S2PEZ7 A0A0J7KLH8 A0A0J7KKI5 X1WPJ7 A0A0J7NFZ0 J9KNG8
Q8MY31 Q8MY33 Q8MY35 A0A2W1C3J5 A0A0J7KCI1 O01419 A0A0J7N1D9 A0A0J7N7H9 A0A0J7KCQ3 A0A0J7MY07 A0A0J7MZM4 A0A0J7KS67 A0A0J7KJB9 E2BVR6 A0A0J7KM11 A0A194R5D3 A0A0J7KIY3 A0A0J7KJH4 A0A0J7KIY5 A0A2H1WNZ5 A0A0J7KLT9 A0A0J7JZF7 A0A0J7JZ13 A0A0L7R7C3 A0A0J7KJG6 A0A0J7N7S8 A0A0J7JXN3 A0A0J7NEG9 A0A0J7KQK3 A0A3L8DIL1 A0A3L8DDV9 A0A0J7MX22 A0A3L8D2G3 A0A0J7KFG9 A0A0J7KIW1 A0A2S2NPF9 A0A0J7KIM6 A0A2S2PJ67 A0A0J7KPR0 A0A0J7K3F1 A0A0J7KYD2 J9KRD0 A0A0J7K840 A0A0J7KWU5 A0A0J7JYP2 A0A2S2P6S8 A0A0J7KPZ3 J9KJG8 A0A0J7KRB8 E2A7V5 A0A0J7NCZ4 A0A0J7NMN5 A0A0J7NAV8 A0A0J7KL06 A0A2H8TR36 A0A0J7KC11 A0A2S2NME4 A0A2S2PYG9 J9KLZ3 A0A2S2PSC1 A0A2S2NJ82 A0A3Q0JIB3 X1WJY4 A0A0J7K6Z4 A0A0J7KRU6 A0A0Q9WRU0 A0A0J7MY17 A0A2S2PEU2 J9LAC1 A0A2H8THF8 J9LDW8 A0A2S2PDK9 A0A0J7KIA8 A0A0J7KF40 J9LJ80 A0A2S2PEZ7 A0A0J7KLH8 A0A0J7KKI5 X1WPJ7 A0A0J7NFZ0 J9KNG8
Ontologies
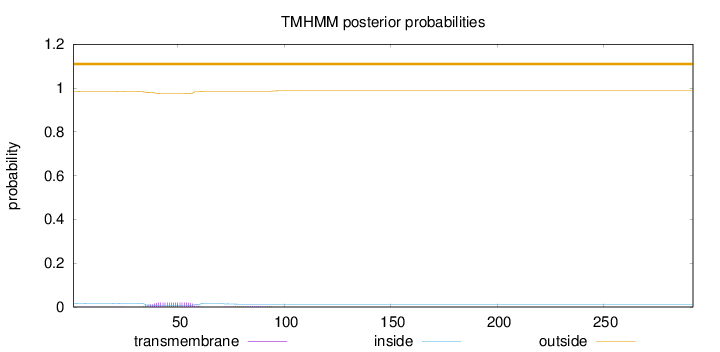
Topology
Length:
292
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.531189999999999
Exp number, first 60 AAs:
0.46053
Total prob of N-in:
0.01596
outside
1 - 292
Population Genetic Test Statistics
Pi
19.396148
Theta
21.128503
Tajima's D
-0.210569
CLR
0
CSRT
0.315184240787961
Interpretation
Uncertain
