Gene
KWMTBOMO00120
Pre Gene Modal
BGIBMGA002134
Annotation
Band_7_protein_AAEL010189_[Papilio_xuthus]
Full name
Band 7 protein AAEL010189
+ More
Band 7 protein AGAP004871
Band 7 protein CG42540
Band 7 protein AGAP004871
Band 7 protein CG42540
Location in the cell
PlasmaMembrane Reliability : 1.401
Sequence
CDS
ATGGTGGTGACCGAAGCGACAGCGAATGAAGACGTCGACTCAGAGTCGAAGACATGTGGCAAGATCCTGGTGGTGCTATCATGGATGCTGGTGATCCTCACCATGCCGTTTTCGCTCTTCATATGCTTCAAGGTGGTGCAAGAATATGAAAGAGCTGTAATTTTTCGCTTGGGGCGTTTACTGTCTGGAGGTGCCAAAGGACCCGGAATATTCTTCATCCTGCCCTGCATTGACACGTACGCCCGAGTCGACCTACGCACACGTACCTATGATGTTCCACCACAAGAAGTGCTGACCAAAGATTCCGTAACTGTTTCCGTGGACGCCGTCGTATACTACCGCGTCCACAACGCGACTATCTCTATCGCCAACGTGGAGAACGCTCACCACTCCACCAGGCTCTTGGCTCAGACGACCTTGAGGAATACAATGGGCACTCGACCTCTGCACGAGATCTTGAGTGAGCGCGAAACGATCTCTGGCAACATGCAGCTGTCACTGGATGAGGCCACCGAGGCTTGGGGGATCAAAGTGGAGCGTGTTGAGATCAAAGACGTCCGTCTACCGGTGCAGTTGCAACGTGCCATGGCCGCCGAAGCCGAGGCTGCTCGCGAGGCTCGTGCCAAGGTCATCGCCGCCGAGGGAGAACAGAAAGCTTCTCGAGCTCTCCGTGAGGCCTCAGAAGTCATTGGTGACAGCCCTGCCGCTTTACAGCTACGATACCTACAGACCCTCAATACAATTTCGGCTGAGAAGAACTCGACCATCGTGTTTCCACTGCCAATCGACCTCCTCACCTACTTCATCAAGGCGCGAGAGGAACCCGTGTAA
Protein
MVVTEATANEDVDSESKTCGKILVVLSWMLVILTMPFSLFICFKVVQEYERAVIFRLGRLLSGGAKGPGIFFILPCIDTYARVDLRTRTYDVPPQEVLTKDSVTVSVDAVVYYRVHNATISIANVENAHHSTRLLAQTTLRNTMGTRPLHEILSERETISGNMQLSLDEATEAWGIKVERVEIKDVRLPVQLQRAMAAEAEAAREARAKVIAAEGEQKASRALREASEVIGDSPAALQLRYLQTLNTISAEKNSTIVFPLPIDLLTYFIKAREEPV
Summary
Similarity
Belongs to the band 7/mec-2 family.
Keywords
Complete proteome
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Alternative splicing
RNA editing
Feature
chain Band 7 protein AAEL010189
splice variant In isoform F.
splice variant In isoform F.
Uniprot
A0A212F9T4
A0A2A4JK66
A0A2H1WMJ4
A0A194QZT4
A0A194Q4U9
K7ILS8
+ More
A0A232F9E7 A0A195C3L6 E2C3C8 E2A6U3 B0XH59 E9IPG1 A0A087ZS70 A0A151X7S6 A0A182J6Q3 A0A2A3ENB7 A0A182QE00 A0A182PSX9 A0A182FDY6 Q16TM5 A0A310SQ57 Q7PPU9 A0A182XLD2 A0A182HXR7 A0A0M9A854 D6WI38 W5J3R3 A0A1W4WZY1 A0A336LJ09 U4TYQ6 A0A336LMH2 A0A0Q9WAV2 A0A0Q9WD75 A0A0L7RCN1 A0A0R3P685 A0A1W4VAX2 A0A0P8XSU2 A0A0A1WQD4 B4MFY5 B4IZ81 A0A1W4VNB4 A0A0J9RPJ8 A0A0R3P8X7 Q9VZA4-2 A0A0Q5U795 A0A0P8Y8M3 A0A2J7QPG8 A0A0R3P8N3 A0A0J9RQK3 A0A0R1DVV7 Q9VZA4-3 A0A2J7QPF9 A0A1W4VNZ5 A0A1W4VMK9 A0A0P8YFU8 B4QR04 B4HU85 A0A1W4VAK3 A0A0N8P0Q7 A0A0L0C6W0 A0A0Q9W8G8 W8AN51 A0A0P8XT88 A0A0L0C6Y6 A0A0J9RQJ9 A0A2J7QPF8 A0A0J9RPJ4 M9PEM4 X2JC20 A0A0Q5U3C3 B3NC60 A0A0R1DVW1 B5DRC5 A0A0R1E102 N6TP25 A0A1W4VNZ1 A0A0J9RP50 A0A182M3L5 A0A182V8L1 A0A182YBT5 M9PEB7 A0A0P8XSU7 A0A034WI56 A0A0Q9WPH7 A0A0Q9VZV9 A0A0Q5UEI7 B4KX97 A0A0R3P6A6 B3M9I3 A0A0J9RNX8 H8F4R7 Q9VZA4 W8B2D5 A0A0P9AIY3 A0A0K8VAU6 A0A0R3P6A1 A0A0R1DVQ5 A0A0L0C9B1 A0A1W4VAW7 A0A1A9UXN8
A0A232F9E7 A0A195C3L6 E2C3C8 E2A6U3 B0XH59 E9IPG1 A0A087ZS70 A0A151X7S6 A0A182J6Q3 A0A2A3ENB7 A0A182QE00 A0A182PSX9 A0A182FDY6 Q16TM5 A0A310SQ57 Q7PPU9 A0A182XLD2 A0A182HXR7 A0A0M9A854 D6WI38 W5J3R3 A0A1W4WZY1 A0A336LJ09 U4TYQ6 A0A336LMH2 A0A0Q9WAV2 A0A0Q9WD75 A0A0L7RCN1 A0A0R3P685 A0A1W4VAX2 A0A0P8XSU2 A0A0A1WQD4 B4MFY5 B4IZ81 A0A1W4VNB4 A0A0J9RPJ8 A0A0R3P8X7 Q9VZA4-2 A0A0Q5U795 A0A0P8Y8M3 A0A2J7QPG8 A0A0R3P8N3 A0A0J9RQK3 A0A0R1DVV7 Q9VZA4-3 A0A2J7QPF9 A0A1W4VNZ5 A0A1W4VMK9 A0A0P8YFU8 B4QR04 B4HU85 A0A1W4VAK3 A0A0N8P0Q7 A0A0L0C6W0 A0A0Q9W8G8 W8AN51 A0A0P8XT88 A0A0L0C6Y6 A0A0J9RQJ9 A0A2J7QPF8 A0A0J9RPJ4 M9PEM4 X2JC20 A0A0Q5U3C3 B3NC60 A0A0R1DVW1 B5DRC5 A0A0R1E102 N6TP25 A0A1W4VNZ1 A0A0J9RP50 A0A182M3L5 A0A182V8L1 A0A182YBT5 M9PEB7 A0A0P8XSU7 A0A034WI56 A0A0Q9WPH7 A0A0Q9VZV9 A0A0Q5UEI7 B4KX97 A0A0R3P6A6 B3M9I3 A0A0J9RNX8 H8F4R7 Q9VZA4 W8B2D5 A0A0P9AIY3 A0A0K8VAU6 A0A0R3P6A1 A0A0R1DVQ5 A0A0L0C9B1 A0A1W4VAW7 A0A1A9UXN8
Pubmed
EMBL
AGBW02009564
OWR50483.1
NWSH01001185
PCG72219.1
ODYU01009690
SOQ54301.1
+ More
KQ461108 KPJ09101.1 KQ459463 KPJ00567.1 NNAY01000670 OXU27079.1 KQ978344 KYM94781.1 GL452320 EFN77466.1 GL437203 EFN70809.1 DS233104 EDS28121.1 GL764503 EFZ17549.1 KQ982431 KYQ56432.1 KZ288204 PBC33190.1 AXCN02001918 CH477644 EAT37869.1 KQ760382 OAD60709.1 AAAB01008905 EAA09720.4 APCN01005337 KQ435714 KOX79200.1 KQ971334 EFA01055.2 ADMH02002184 ETN57948.1 UFQT01000016 SSX17830.1 KB631855 ERL86759.1 SSX17829.1 CH940669 KRF78191.1 KRF78192.1 KQ414615 KOC68747.1 CH379070 KRT08673.1 CH902618 KPU77798.1 GBXI01013476 JAD00816.1 EDW58246.2 CH916366 EDV95603.1 CM002912 KMY97647.1 KRT08675.1 AE014296 AY060624 AY069196 BT021331 AAF47920.2 AAL28172.1 AAX33479.1 CH954178 KQS43604.1 KPU77797.1 NEVH01012091 PNF30471.1 KRT08677.1 KMY97644.1 CM000159 KRK01197.1 PNF30472.1 KPU77802.1 CM000363 EDX09241.1 CH480817 EDW50506.1 KPU77796.1 JRES01000828 KNC28005.1 KRF78193.1 GAMC01019136 GAMC01019133 JAB87422.1 KPU77801.1 KNC28001.1 KMY97639.1 PNF30475.1 KMY97642.1 AGB94114.1 AHN57961.1 KQS43599.1 EDV50948.2 KRK01193.1 EDY74047.2 KRK01194.1 APGK01057589 APGK01057590 APGK01057591 APGK01057592 APGK01057593 APGK01057594 APGK01057595 KB741280 ENN70980.1 KMY97643.1 AXCM01009917 AXCM01009918 AGB94115.1 KPU77800.1 GAKP01004588 JAC54364.1 CH963847 KRF97782.1 KRF78194.1 KQS43601.1 CH933809 EDW17555.2 KRT08676.1 EDV38989.2 KMY97641.1 BT133307 AFD10754.1 GAMC01019137 GAMC01019134 JAB87421.1 KPU77799.1 GDHF01027774 GDHF01016342 JAI24540.1 JAI35972.1 KRT08674.1 KRK01196.1 KNC28004.1
KQ461108 KPJ09101.1 KQ459463 KPJ00567.1 NNAY01000670 OXU27079.1 KQ978344 KYM94781.1 GL452320 EFN77466.1 GL437203 EFN70809.1 DS233104 EDS28121.1 GL764503 EFZ17549.1 KQ982431 KYQ56432.1 KZ288204 PBC33190.1 AXCN02001918 CH477644 EAT37869.1 KQ760382 OAD60709.1 AAAB01008905 EAA09720.4 APCN01005337 KQ435714 KOX79200.1 KQ971334 EFA01055.2 ADMH02002184 ETN57948.1 UFQT01000016 SSX17830.1 KB631855 ERL86759.1 SSX17829.1 CH940669 KRF78191.1 KRF78192.1 KQ414615 KOC68747.1 CH379070 KRT08673.1 CH902618 KPU77798.1 GBXI01013476 JAD00816.1 EDW58246.2 CH916366 EDV95603.1 CM002912 KMY97647.1 KRT08675.1 AE014296 AY060624 AY069196 BT021331 AAF47920.2 AAL28172.1 AAX33479.1 CH954178 KQS43604.1 KPU77797.1 NEVH01012091 PNF30471.1 KRT08677.1 KMY97644.1 CM000159 KRK01197.1 PNF30472.1 KPU77802.1 CM000363 EDX09241.1 CH480817 EDW50506.1 KPU77796.1 JRES01000828 KNC28005.1 KRF78193.1 GAMC01019136 GAMC01019133 JAB87422.1 KPU77801.1 KNC28001.1 KMY97639.1 PNF30475.1 KMY97642.1 AGB94114.1 AHN57961.1 KQS43599.1 EDV50948.2 KRK01193.1 EDY74047.2 KRK01194.1 APGK01057589 APGK01057590 APGK01057591 APGK01057592 APGK01057593 APGK01057594 APGK01057595 KB741280 ENN70980.1 KMY97643.1 AXCM01009917 AXCM01009918 AGB94115.1 KPU77800.1 GAKP01004588 JAC54364.1 CH963847 KRF97782.1 KRF78194.1 KQS43601.1 CH933809 EDW17555.2 KRT08676.1 EDV38989.2 KMY97641.1 BT133307 AFD10754.1 GAMC01019137 GAMC01019134 JAB87421.1 KPU77799.1 GDHF01027774 GDHF01016342 JAI24540.1 JAI35972.1 KRT08674.1 KRK01196.1 KNC28004.1
Proteomes
UP000007151
UP000218220
UP000053240
UP000053268
UP000002358
UP000215335
+ More
UP000078542 UP000008237 UP000000311 UP000002320 UP000005203 UP000075809 UP000075880 UP000242457 UP000075886 UP000075885 UP000069272 UP000008820 UP000007062 UP000076407 UP000075840 UP000053105 UP000007266 UP000000673 UP000192223 UP000030742 UP000008792 UP000053825 UP000001819 UP000192221 UP000007801 UP000001070 UP000000803 UP000008711 UP000235965 UP000002282 UP000000304 UP000001292 UP000037069 UP000019118 UP000075883 UP000075903 UP000076408 UP000007798 UP000009192 UP000078200
UP000078542 UP000008237 UP000000311 UP000002320 UP000005203 UP000075809 UP000075880 UP000242457 UP000075886 UP000075885 UP000069272 UP000008820 UP000007062 UP000076407 UP000075840 UP000053105 UP000007266 UP000000673 UP000192223 UP000030742 UP000008792 UP000053825 UP000001819 UP000192221 UP000007801 UP000001070 UP000000803 UP000008711 UP000235965 UP000002282 UP000000304 UP000001292 UP000037069 UP000019118 UP000075883 UP000075903 UP000076408 UP000007798 UP000009192 UP000078200
Pfam
PF01145 Band_7
Interpro
Gene 3D
ProteinModelPortal
A0A212F9T4
A0A2A4JK66
A0A2H1WMJ4
A0A194QZT4
A0A194Q4U9
K7ILS8
+ More
A0A232F9E7 A0A195C3L6 E2C3C8 E2A6U3 B0XH59 E9IPG1 A0A087ZS70 A0A151X7S6 A0A182J6Q3 A0A2A3ENB7 A0A182QE00 A0A182PSX9 A0A182FDY6 Q16TM5 A0A310SQ57 Q7PPU9 A0A182XLD2 A0A182HXR7 A0A0M9A854 D6WI38 W5J3R3 A0A1W4WZY1 A0A336LJ09 U4TYQ6 A0A336LMH2 A0A0Q9WAV2 A0A0Q9WD75 A0A0L7RCN1 A0A0R3P685 A0A1W4VAX2 A0A0P8XSU2 A0A0A1WQD4 B4MFY5 B4IZ81 A0A1W4VNB4 A0A0J9RPJ8 A0A0R3P8X7 Q9VZA4-2 A0A0Q5U795 A0A0P8Y8M3 A0A2J7QPG8 A0A0R3P8N3 A0A0J9RQK3 A0A0R1DVV7 Q9VZA4-3 A0A2J7QPF9 A0A1W4VNZ5 A0A1W4VMK9 A0A0P8YFU8 B4QR04 B4HU85 A0A1W4VAK3 A0A0N8P0Q7 A0A0L0C6W0 A0A0Q9W8G8 W8AN51 A0A0P8XT88 A0A0L0C6Y6 A0A0J9RQJ9 A0A2J7QPF8 A0A0J9RPJ4 M9PEM4 X2JC20 A0A0Q5U3C3 B3NC60 A0A0R1DVW1 B5DRC5 A0A0R1E102 N6TP25 A0A1W4VNZ1 A0A0J9RP50 A0A182M3L5 A0A182V8L1 A0A182YBT5 M9PEB7 A0A0P8XSU7 A0A034WI56 A0A0Q9WPH7 A0A0Q9VZV9 A0A0Q5UEI7 B4KX97 A0A0R3P6A6 B3M9I3 A0A0J9RNX8 H8F4R7 Q9VZA4 W8B2D5 A0A0P9AIY3 A0A0K8VAU6 A0A0R3P6A1 A0A0R1DVQ5 A0A0L0C9B1 A0A1W4VAW7 A0A1A9UXN8
A0A232F9E7 A0A195C3L6 E2C3C8 E2A6U3 B0XH59 E9IPG1 A0A087ZS70 A0A151X7S6 A0A182J6Q3 A0A2A3ENB7 A0A182QE00 A0A182PSX9 A0A182FDY6 Q16TM5 A0A310SQ57 Q7PPU9 A0A182XLD2 A0A182HXR7 A0A0M9A854 D6WI38 W5J3R3 A0A1W4WZY1 A0A336LJ09 U4TYQ6 A0A336LMH2 A0A0Q9WAV2 A0A0Q9WD75 A0A0L7RCN1 A0A0R3P685 A0A1W4VAX2 A0A0P8XSU2 A0A0A1WQD4 B4MFY5 B4IZ81 A0A1W4VNB4 A0A0J9RPJ8 A0A0R3P8X7 Q9VZA4-2 A0A0Q5U795 A0A0P8Y8M3 A0A2J7QPG8 A0A0R3P8N3 A0A0J9RQK3 A0A0R1DVV7 Q9VZA4-3 A0A2J7QPF9 A0A1W4VNZ5 A0A1W4VMK9 A0A0P8YFU8 B4QR04 B4HU85 A0A1W4VAK3 A0A0N8P0Q7 A0A0L0C6W0 A0A0Q9W8G8 W8AN51 A0A0P8XT88 A0A0L0C6Y6 A0A0J9RQJ9 A0A2J7QPF8 A0A0J9RPJ4 M9PEM4 X2JC20 A0A0Q5U3C3 B3NC60 A0A0R1DVW1 B5DRC5 A0A0R1E102 N6TP25 A0A1W4VNZ1 A0A0J9RP50 A0A182M3L5 A0A182V8L1 A0A182YBT5 M9PEB7 A0A0P8XSU7 A0A034WI56 A0A0Q9WPH7 A0A0Q9VZV9 A0A0Q5UEI7 B4KX97 A0A0R3P6A6 B3M9I3 A0A0J9RNX8 H8F4R7 Q9VZA4 W8B2D5 A0A0P9AIY3 A0A0K8VAU6 A0A0R3P6A1 A0A0R1DVQ5 A0A0L0C9B1 A0A1W4VAW7 A0A1A9UXN8
PDB
4FVF
E-value=3.40018e-41,
Score=421
Ontologies
KEGG
GO
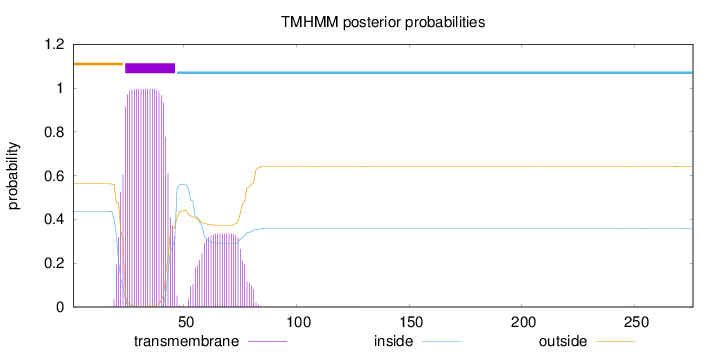
Topology
Subcellular location
Membrane
Length:
276
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
29.3671
Exp number, first 60 AAs:
23.59099
Total prob of N-in:
0.43729
POSSIBLE N-term signal
sequence
outside
1 - 23
TMhelix
24 - 46
inside
47 - 276
Population Genetic Test Statistics
Pi
18.614041
Theta
19.975758
Tajima's D
-1.106489
CLR
0.085998
CSRT
0.117994100294985
Interpretation
Uncertain
