Gene
KWMTBOMO00119
Pre Gene Modal
BGIBMGA002133
Annotation
PREDICTED:_uncharacterized_protein_LOC101744434_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.972
Sequence
CDS
ATGATAACAGCTGACGAAGGCTGCCTCCAGTACTACACGGGCGTGTCTGGTCAATTACGCTCTTTCAACTATGACCCAGCCTCAGGATTCCAGCTGAGTAATCAAGACTACGGCATCTGCATCAGAATGGAGAGGAACTTCTGCGGTATCCTGTACACCGCCTGCCCCGACCAAGGTATATTTAAAATTAAAAGTCTTTAA
Protein
MITADEGCLQYYTGVSGQLRSFNYDPASGFQLSNQDYGICIRMERNFCGILYTACPDQGIFKIKSL
Summary
Uniprot
H9IY00
A0A194Q4U9
A0A2H1WMK1
A0A2A4J7F9
A0A194QVV9
A0A0L7L2D1
+ More
A0A067QY02 A0A2P8Z0Y0 A0A1B0CVV7 A0A2R7X2G9 A0A182YBT4 A0A212F9R8 E9IPG0 E2A6U4 T1HWE2 A0A1B6HUK6 A0A026W2S8 A0A3Q0J4Q0 A0A2J7QPF6 E2C3C9 A0A2J7QPG0 A0A3L8D5J0 A0A195C492 F4WBE3 A0A151JYU5 A0A151X9V1 A0A0N0U7B5 A0A151JNN1 A0A2A3EQ77 A0A087ZSA5 A0A151HXZ9 A0A1B6E7L8 A0A067QW23 A0A182IYE2 A0A182STN0 A0A154PJ35 W5J5C3 A0A139WK06 A0A139WKC0 A0A0L0CLD6 A0A084WJB0 A0A182NNK4 A0A1I8M8F7 A0A182FDY7 K7J6G6 A0A182TJR3 A0A182VCJ6 A0A182XLD3 Q7Q975 A0A182HXR6 A0A182LHY7 A0A0K2T1W4 J9JQT2 E0W1S6 A0A182Q8D7 A0A182PSY0 A0A1Y1LBX5 A0A182WMM0 A0A182MUV0 A0A182K3A7 Q172Q1 A0A1S4FPS2 A0A182RKI0 A0A310SSU1 Q16TM6 A0A1I8ML27 A0A182GPS9 A0A0L7RD12 A0A1I8NSI1 B4MMD9 B4LEN9 A0A1W4VLD9 B3NBN3 B4PFI8 A0A0M4EMH5 B4HK30 Q9VSQ2 B4QM79 A0A1A9W3G3 B3M5Y9 A0A1A9XK55 B4KZF4 A0A1J1ISJ9 A0A1B0ANQ9 A0A1B0FBQ5 A0A2S2PER1 A0A1A9UP65 U4U9X9 N6STG9 B4J3M4 Q29D25 B4H9L3 A0A3B0J6F9 A0A336K0W9 A0A2S2NC08 A0A226EKW6 T1GGB9 A0A1D2MTG5 A0A226ELE8 A0A034UYC1
A0A067QY02 A0A2P8Z0Y0 A0A1B0CVV7 A0A2R7X2G9 A0A182YBT4 A0A212F9R8 E9IPG0 E2A6U4 T1HWE2 A0A1B6HUK6 A0A026W2S8 A0A3Q0J4Q0 A0A2J7QPF6 E2C3C9 A0A2J7QPG0 A0A3L8D5J0 A0A195C492 F4WBE3 A0A151JYU5 A0A151X9V1 A0A0N0U7B5 A0A151JNN1 A0A2A3EQ77 A0A087ZSA5 A0A151HXZ9 A0A1B6E7L8 A0A067QW23 A0A182IYE2 A0A182STN0 A0A154PJ35 W5J5C3 A0A139WK06 A0A139WKC0 A0A0L0CLD6 A0A084WJB0 A0A182NNK4 A0A1I8M8F7 A0A182FDY7 K7J6G6 A0A182TJR3 A0A182VCJ6 A0A182XLD3 Q7Q975 A0A182HXR6 A0A182LHY7 A0A0K2T1W4 J9JQT2 E0W1S6 A0A182Q8D7 A0A182PSY0 A0A1Y1LBX5 A0A182WMM0 A0A182MUV0 A0A182K3A7 Q172Q1 A0A1S4FPS2 A0A182RKI0 A0A310SSU1 Q16TM6 A0A1I8ML27 A0A182GPS9 A0A0L7RD12 A0A1I8NSI1 B4MMD9 B4LEN9 A0A1W4VLD9 B3NBN3 B4PFI8 A0A0M4EMH5 B4HK30 Q9VSQ2 B4QM79 A0A1A9W3G3 B3M5Y9 A0A1A9XK55 B4KZF4 A0A1J1ISJ9 A0A1B0ANQ9 A0A1B0FBQ5 A0A2S2PER1 A0A1A9UP65 U4U9X9 N6STG9 B4J3M4 Q29D25 B4H9L3 A0A3B0J6F9 A0A336K0W9 A0A2S2NC08 A0A226EKW6 T1GGB9 A0A1D2MTG5 A0A226ELE8 A0A034UYC1
Pubmed
19121390
26354079
26227816
24845553
29403074
25244985
+ More
22118469 21282665 20798317 24508170 30249741 21719571 20920257 23761445 18362917 19820115 26108605 24438588 25315136 20075255 12364791 14747013 17210077 20966253 20566863 28004739 17510324 26483478 17994087 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 23537049 15632085 27289101 25348373
22118469 21282665 20798317 24508170 30249741 21719571 20920257 23761445 18362917 19820115 26108605 24438588 25315136 20075255 12364791 14747013 17210077 20966253 20566863 28004739 17510324 26483478 17994087 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 23537049 15632085 27289101 25348373
EMBL
BABH01015913
KQ459463
KPJ00567.1
ODYU01009690
SOQ54300.1
NWSH01002531
+ More
PCG68057.1 KQ461108 KPJ09100.1 JTDY01003421 KOB69580.1 KK852891 KDR14261.1 PYGN01000250 PSN50138.1 AJWK01031317 AJWK01031318 AJWK01031319 AJWK01031320 AJWK01031321 AJWK01031322 AJWK01031323 AJWK01031324 AJWK01031325 AJWK01031326 KK856359 PTY25580.1 AGBW02009564 OWR50482.1 GL764503 EFZ17537.1 GL437203 EFN70810.1 ACPB03014114 GECU01029342 JAS78364.1 KK107462 EZA50318.1 NEVH01012091 PNF30468.1 GL452320 EFN77467.1 PNF30477.1 QOIP01000013 RLU15494.1 KQ978344 KYM94988.1 GL888062 EGI68499.1 KQ981451 KYN41622.1 KQ982365 KYQ57114.1 KQ435714 KOX79201.1 KQ978820 KYN28121.1 KZ288204 PBC33191.1 KQ976739 KYM75618.1 GEDC01003378 JAS33920.1 KDR14260.1 KQ434931 KZC11855.1 ADMH02002184 ETN57949.1 KQ971334 KYB28256.1 KYB28257.1 JRES01000234 KNC33110.1 ATLV01024006 KE525348 KFB50304.1 AAAB01008905 EAA09694.1 APCN01005337 HACA01002444 CDW19805.1 ABLF02024083 ABLF02024220 ABLF02041348 DS235873 EEB19658.1 AXCN02001918 GEZM01060273 JAV71134.1 AXCM01009919 CH477433 EAT40997.1 KQ760382 OAD60708.1 CH477644 EAT37868.1 JXUM01079029 JXUM01079030 KQ563103 KXJ74455.1 KQ414615 KOC68748.1 CH963847 EDW73284.2 CH940647 EDW69124.1 KRF84192.1 CH954178 EDV50629.1 CM000159 EDW93114.1 CP012525 ALC44885.1 CH480815 EDW40766.1 AE014296 AAF50364.1 CM000363 CM002912 EDX09762.1 KMY98479.1 CH902618 EDV39679.2 CH933809 EDW17881.1 CVRI01000059 CRL03175.1 JXJN01000915 CCAG010004106 GGMR01015069 MBY27688.1 KB631855 ERL86760.1 APGK01057588 KB741280 ENN70979.1 CH916366 EDV96226.1 CH379070 EAL30589.2 CH479229 EDW36489.1 OUUW01000002 SPP77365.1 UFQS01000016 UFQT01000016 SSW97442.1 SSX17828.1 GGMR01001863 MBY14482.1 LNIX01000003 OXA57226.1 CAQQ02023106 LJIJ01000549 ODM96333.1 OXA57396.1 GAKP01022958 JAC35996.1
PCG68057.1 KQ461108 KPJ09100.1 JTDY01003421 KOB69580.1 KK852891 KDR14261.1 PYGN01000250 PSN50138.1 AJWK01031317 AJWK01031318 AJWK01031319 AJWK01031320 AJWK01031321 AJWK01031322 AJWK01031323 AJWK01031324 AJWK01031325 AJWK01031326 KK856359 PTY25580.1 AGBW02009564 OWR50482.1 GL764503 EFZ17537.1 GL437203 EFN70810.1 ACPB03014114 GECU01029342 JAS78364.1 KK107462 EZA50318.1 NEVH01012091 PNF30468.1 GL452320 EFN77467.1 PNF30477.1 QOIP01000013 RLU15494.1 KQ978344 KYM94988.1 GL888062 EGI68499.1 KQ981451 KYN41622.1 KQ982365 KYQ57114.1 KQ435714 KOX79201.1 KQ978820 KYN28121.1 KZ288204 PBC33191.1 KQ976739 KYM75618.1 GEDC01003378 JAS33920.1 KDR14260.1 KQ434931 KZC11855.1 ADMH02002184 ETN57949.1 KQ971334 KYB28256.1 KYB28257.1 JRES01000234 KNC33110.1 ATLV01024006 KE525348 KFB50304.1 AAAB01008905 EAA09694.1 APCN01005337 HACA01002444 CDW19805.1 ABLF02024083 ABLF02024220 ABLF02041348 DS235873 EEB19658.1 AXCN02001918 GEZM01060273 JAV71134.1 AXCM01009919 CH477433 EAT40997.1 KQ760382 OAD60708.1 CH477644 EAT37868.1 JXUM01079029 JXUM01079030 KQ563103 KXJ74455.1 KQ414615 KOC68748.1 CH963847 EDW73284.2 CH940647 EDW69124.1 KRF84192.1 CH954178 EDV50629.1 CM000159 EDW93114.1 CP012525 ALC44885.1 CH480815 EDW40766.1 AE014296 AAF50364.1 CM000363 CM002912 EDX09762.1 KMY98479.1 CH902618 EDV39679.2 CH933809 EDW17881.1 CVRI01000059 CRL03175.1 JXJN01000915 CCAG010004106 GGMR01015069 MBY27688.1 KB631855 ERL86760.1 APGK01057588 KB741280 ENN70979.1 CH916366 EDV96226.1 CH379070 EAL30589.2 CH479229 EDW36489.1 OUUW01000002 SPP77365.1 UFQS01000016 UFQT01000016 SSW97442.1 SSX17828.1 GGMR01001863 MBY14482.1 LNIX01000003 OXA57226.1 CAQQ02023106 LJIJ01000549 ODM96333.1 OXA57396.1 GAKP01022958 JAC35996.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000037510
UP000027135
+ More
UP000245037 UP000092461 UP000076408 UP000007151 UP000000311 UP000015103 UP000053097 UP000079169 UP000235965 UP000008237 UP000279307 UP000078542 UP000007755 UP000078541 UP000075809 UP000053105 UP000078492 UP000242457 UP000005203 UP000078540 UP000075880 UP000075901 UP000076502 UP000000673 UP000007266 UP000037069 UP000030765 UP000075884 UP000095301 UP000069272 UP000002358 UP000075902 UP000075903 UP000076407 UP000007062 UP000075840 UP000075882 UP000007819 UP000009046 UP000075886 UP000075885 UP000075920 UP000075883 UP000075881 UP000008820 UP000075900 UP000069940 UP000249989 UP000053825 UP000095300 UP000007798 UP000008792 UP000192221 UP000008711 UP000002282 UP000092553 UP000001292 UP000000803 UP000000304 UP000091820 UP000007801 UP000092443 UP000009192 UP000183832 UP000092460 UP000092444 UP000078200 UP000030742 UP000019118 UP000001070 UP000001819 UP000008744 UP000268350 UP000198287 UP000015102 UP000094527
UP000245037 UP000092461 UP000076408 UP000007151 UP000000311 UP000015103 UP000053097 UP000079169 UP000235965 UP000008237 UP000279307 UP000078542 UP000007755 UP000078541 UP000075809 UP000053105 UP000078492 UP000242457 UP000005203 UP000078540 UP000075880 UP000075901 UP000076502 UP000000673 UP000007266 UP000037069 UP000030765 UP000075884 UP000095301 UP000069272 UP000002358 UP000075902 UP000075903 UP000076407 UP000007062 UP000075840 UP000075882 UP000007819 UP000009046 UP000075886 UP000075885 UP000075920 UP000075883 UP000075881 UP000008820 UP000075900 UP000069940 UP000249989 UP000053825 UP000095300 UP000007798 UP000008792 UP000192221 UP000008711 UP000002282 UP000092553 UP000001292 UP000000803 UP000000304 UP000091820 UP000007801 UP000092443 UP000009192 UP000183832 UP000092460 UP000092444 UP000078200 UP000030742 UP000019118 UP000001070 UP000001819 UP000008744 UP000268350 UP000198287 UP000015102 UP000094527
Interpro
IPR001107
Band_7
+ More
IPR018080 Band_7/stomatin-like_CS
IPR036013 Band_7/SPFH_dom_sf
IPR001972 Stomatin_fam
IPR000859 CUB_dom
IPR035914 Sperma_CUB_dom_sf
IPR029021 Prot-tyrosine_phosphatase-like
IPR015425 FH2_Formin
IPR014767 DAD_dom
IPR011989 ARM-like
IPR016024 ARM-type_fold
IPR014768 GBD/FH3_dom
IPR000477 RT_dom
IPR018080 Band_7/stomatin-like_CS
IPR036013 Band_7/SPFH_dom_sf
IPR001972 Stomatin_fam
IPR000859 CUB_dom
IPR035914 Sperma_CUB_dom_sf
IPR029021 Prot-tyrosine_phosphatase-like
IPR015425 FH2_Formin
IPR014767 DAD_dom
IPR011989 ARM-like
IPR016024 ARM-type_fold
IPR014768 GBD/FH3_dom
IPR000477 RT_dom
Gene 3D
ProteinModelPortal
H9IY00
A0A194Q4U9
A0A2H1WMK1
A0A2A4J7F9
A0A194QVV9
A0A0L7L2D1
+ More
A0A067QY02 A0A2P8Z0Y0 A0A1B0CVV7 A0A2R7X2G9 A0A182YBT4 A0A212F9R8 E9IPG0 E2A6U4 T1HWE2 A0A1B6HUK6 A0A026W2S8 A0A3Q0J4Q0 A0A2J7QPF6 E2C3C9 A0A2J7QPG0 A0A3L8D5J0 A0A195C492 F4WBE3 A0A151JYU5 A0A151X9V1 A0A0N0U7B5 A0A151JNN1 A0A2A3EQ77 A0A087ZSA5 A0A151HXZ9 A0A1B6E7L8 A0A067QW23 A0A182IYE2 A0A182STN0 A0A154PJ35 W5J5C3 A0A139WK06 A0A139WKC0 A0A0L0CLD6 A0A084WJB0 A0A182NNK4 A0A1I8M8F7 A0A182FDY7 K7J6G6 A0A182TJR3 A0A182VCJ6 A0A182XLD3 Q7Q975 A0A182HXR6 A0A182LHY7 A0A0K2T1W4 J9JQT2 E0W1S6 A0A182Q8D7 A0A182PSY0 A0A1Y1LBX5 A0A182WMM0 A0A182MUV0 A0A182K3A7 Q172Q1 A0A1S4FPS2 A0A182RKI0 A0A310SSU1 Q16TM6 A0A1I8ML27 A0A182GPS9 A0A0L7RD12 A0A1I8NSI1 B4MMD9 B4LEN9 A0A1W4VLD9 B3NBN3 B4PFI8 A0A0M4EMH5 B4HK30 Q9VSQ2 B4QM79 A0A1A9W3G3 B3M5Y9 A0A1A9XK55 B4KZF4 A0A1J1ISJ9 A0A1B0ANQ9 A0A1B0FBQ5 A0A2S2PER1 A0A1A9UP65 U4U9X9 N6STG9 B4J3M4 Q29D25 B4H9L3 A0A3B0J6F9 A0A336K0W9 A0A2S2NC08 A0A226EKW6 T1GGB9 A0A1D2MTG5 A0A226ELE8 A0A034UYC1
A0A067QY02 A0A2P8Z0Y0 A0A1B0CVV7 A0A2R7X2G9 A0A182YBT4 A0A212F9R8 E9IPG0 E2A6U4 T1HWE2 A0A1B6HUK6 A0A026W2S8 A0A3Q0J4Q0 A0A2J7QPF6 E2C3C9 A0A2J7QPG0 A0A3L8D5J0 A0A195C492 F4WBE3 A0A151JYU5 A0A151X9V1 A0A0N0U7B5 A0A151JNN1 A0A2A3EQ77 A0A087ZSA5 A0A151HXZ9 A0A1B6E7L8 A0A067QW23 A0A182IYE2 A0A182STN0 A0A154PJ35 W5J5C3 A0A139WK06 A0A139WKC0 A0A0L0CLD6 A0A084WJB0 A0A182NNK4 A0A1I8M8F7 A0A182FDY7 K7J6G6 A0A182TJR3 A0A182VCJ6 A0A182XLD3 Q7Q975 A0A182HXR6 A0A182LHY7 A0A0K2T1W4 J9JQT2 E0W1S6 A0A182Q8D7 A0A182PSY0 A0A1Y1LBX5 A0A182WMM0 A0A182MUV0 A0A182K3A7 Q172Q1 A0A1S4FPS2 A0A182RKI0 A0A310SSU1 Q16TM6 A0A1I8ML27 A0A182GPS9 A0A0L7RD12 A0A1I8NSI1 B4MMD9 B4LEN9 A0A1W4VLD9 B3NBN3 B4PFI8 A0A0M4EMH5 B4HK30 Q9VSQ2 B4QM79 A0A1A9W3G3 B3M5Y9 A0A1A9XK55 B4KZF4 A0A1J1ISJ9 A0A1B0ANQ9 A0A1B0FBQ5 A0A2S2PER1 A0A1A9UP65 U4U9X9 N6STG9 B4J3M4 Q29D25 B4H9L3 A0A3B0J6F9 A0A336K0W9 A0A2S2NC08 A0A226EKW6 T1GGB9 A0A1D2MTG5 A0A226ELE8 A0A034UYC1
Ontologies
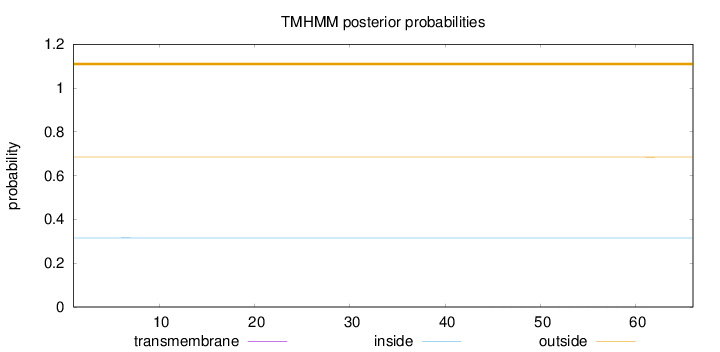
Topology
Length:
66
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00664
Exp number, first 60 AAs:
0.00626
Total prob of N-in:
0.31549
outside
1 - 66
Population Genetic Test Statistics
Pi
17.071796
Theta
16.555493
Tajima's D
0.094143
CLR
0
CSRT
0.400829958502075
Interpretation
Uncertain
