Gene
KWMTBOMO00115
Pre Gene Modal
BGIBMGA002131
Annotation
transient_receptor_potential_cation_channel_subfamily_A_member_1_[Bombyx_mori]
Full name
Transient receptor potential cation channel subfamily A member 1
+ More
Aspartate aminotransferase
Aspartate aminotransferase
Alternative Name
Ankyrin-like with transmembrane domains protein 1
Location in the cell
Cytoplasmic Reliability : 1.25 Extracellular Reliability : 1.199
Sequence
CDS
ATGCTAGCTGTTGGGGCTGCTGAAAGCGGAAATGTCGATGACTTCATGCGATTGTACCTCAGTGAGCCTTCTCGTCTTGCTGTGCGCGATGGAAGGGGTCGTACAGCTGCACATCAAGCCTCTGCTCGAAACAACACTAATATATTGCATTTCATCAATAACTACGCTGGAGACCTTAACGCTAAGGATAACGCCGGGAACACACCGCTCCACGTGGCTGTCGAGAATGAAGCCCTCGACGCTATAGAGTATTTGTTGCAGCAACACGTCGAAACTTCTGTCCTCAACGAGAAGTGTCAAGCTCCAATACACATGGCAACTGAACTAAATAAGGCAAATGTTGGAGAGTCTTAA
Protein
MLAVGAAESGNVDDFMRLYLSEPSRLAVRDGRGRTAAHQASARNNTNILHFINNYAGDLNAKDNAGNTPLHVAVENEALDAIEYLLQQHVETSVLNEKCQAPIHMATELNKANVGES
Summary
Description
Essential for thermotaxis by sensing environmental temperature. Receptor-activated non-selective cation channel involved in detection of sensations such as temperature. Involved in heat nociception by being activated by warm temperature of about 24-29 degrees Celsius.
Catalytic Activity
2-oxoglutarate + L-aspartate = L-glutamate + oxaloacetate
Subunit
Homotetramer.
Homodimer.
Homodimer.
Miscellaneous
In eukaryotes there are cytoplasmic, mitochondrial and chloroplastic isozymes.
Similarity
Belongs to the transient receptor (TC 1.A.4) family.
Keywords
Alternative splicing
ANK repeat
Cell membrane
Complete proteome
Glycoprotein
Ion channel
Ion transport
Membrane
Reference proteome
Repeat
Sensory transduction
Transmembrane
Transmembrane helix
Transport
Feature
chain Transient receptor potential cation channel subfamily A member 1
splice variant In isoform J.
splice variant In isoform J.
Uniprot
H9IXZ8
W8VUV0
X4YI96
A0A2A4J7J5
A0A2A4J8G5
A0A194QAW8
+ More
A0A194QUV6 A0A212F9S1 A0A1A9ZTS1 A0A1Y1M1U7 A0A0Q9X1C4 A0A0Q9WPV3 A0A0Q9WXY6 B4MMD0 A0A0J9RRT9 A0A0J9UHK2 A0A0J9RRT3 Q7Z020-5 H0UST1 Q7Z020 A0A0Q5UHP3 B3NBM3 A0A0Q5U3H8 A0A182HFJ3 B4HK42 A0A0J9RT54 G9L8C4 A0A0J9RS37 H0USS9 A0A0Q5U9M3 A0A0Q5U5B0 M9PBX8 G8IQ59 A0A0M4EIG3 A0A067QW72 A0A0Q9XMU7 A0A0R1DUU3 B4PFH6 A0A0Q9XBB8 A0A0Q9XL17 B4KZG4 U4U429 A0A0P8XTU6 B3M5X8 A0A0P8Y9L5 A0A0P8XTU9 A0A0P9AJX4 A0A0R3P8W6 A0A3B0JXA1 B5DRW8 A0A0R3P941 A0A1W4V0P1 B4LEP7 B4J3L7 A0A2J7RDI1 A0A1B0CK24 D6NLB6 A0A1B0G3C5 A0A2J7RDG6 A0A139WJZ5 A0A1I8PQV0 A0A1I8PQU6 A0A1I8PQU7 A0A1I8PQP5 A0A1S4FME7 A0A1I8NJU2 J9K0X9 A0A1A9V4K4 A0A2S2Q8S0 A0A1W4WSV1 A0A1A9XBH9 A0A1I8QCJ1 A0A1B0BHB3 A0A0R3P7M5 A0A384TFK4 A0A0L0C0A2 A0A1J1HDZ7 A0A182WML1 A0A182HXS5 A0A182WS30 A0A182PV41 A0A182LHX6 A0A182NNL5 A0A182FED1 A0A182M6W7 W5JRE0 A0A182YBU4 A0A182RQE7 B0W5K9 A0A182Q3M9 A0A084W9N1 A0A182UBZ2 Q7Q966 B3G3I7 B0W5K8 E0W214 N6STG4 A0A146KSD5 A0A182JZJ9 A0A182J3R1
A0A194QUV6 A0A212F9S1 A0A1A9ZTS1 A0A1Y1M1U7 A0A0Q9X1C4 A0A0Q9WPV3 A0A0Q9WXY6 B4MMD0 A0A0J9RRT9 A0A0J9UHK2 A0A0J9RRT3 Q7Z020-5 H0UST1 Q7Z020 A0A0Q5UHP3 B3NBM3 A0A0Q5U3H8 A0A182HFJ3 B4HK42 A0A0J9RT54 G9L8C4 A0A0J9RS37 H0USS9 A0A0Q5U9M3 A0A0Q5U5B0 M9PBX8 G8IQ59 A0A0M4EIG3 A0A067QW72 A0A0Q9XMU7 A0A0R1DUU3 B4PFH6 A0A0Q9XBB8 A0A0Q9XL17 B4KZG4 U4U429 A0A0P8XTU6 B3M5X8 A0A0P8Y9L5 A0A0P8XTU9 A0A0P9AJX4 A0A0R3P8W6 A0A3B0JXA1 B5DRW8 A0A0R3P941 A0A1W4V0P1 B4LEP7 B4J3L7 A0A2J7RDI1 A0A1B0CK24 D6NLB6 A0A1B0G3C5 A0A2J7RDG6 A0A139WJZ5 A0A1I8PQV0 A0A1I8PQU6 A0A1I8PQU7 A0A1I8PQP5 A0A1S4FME7 A0A1I8NJU2 J9K0X9 A0A1A9V4K4 A0A2S2Q8S0 A0A1W4WSV1 A0A1A9XBH9 A0A1I8QCJ1 A0A1B0BHB3 A0A0R3P7M5 A0A384TFK4 A0A0L0C0A2 A0A1J1HDZ7 A0A182WML1 A0A182HXS5 A0A182WS30 A0A182PV41 A0A182LHX6 A0A182NNL5 A0A182FED1 A0A182M6W7 W5JRE0 A0A182YBU4 A0A182RQE7 B0W5K9 A0A182Q3M9 A0A084W9N1 A0A182UBZ2 Q7Q966 B3G3I7 B0W5K8 E0W214 N6STG4 A0A146KSD5 A0A182JZJ9 A0A182J3R1
EC Number
2.6.1.1
Pubmed
19121390
24639527
25827167
26354079
22118469
28004739
+ More
17994087 22936249 12815418 10731132 12537572 15681611 26483478 22139422 12537568 12537573 12537574 16110336 17569856 17569867 22347718 26109357 26109356 24845553 17550304 18057021 20237474 23537049 15632085 18362917 19820115 17510324 25315136 26108605 20966253 20920257 23761445 25244985 24438588 12364791 14747013 17210077 18548007 20566863 26823975
17994087 22936249 12815418 10731132 12537572 15681611 26483478 22139422 12537568 12537573 12537574 16110336 17569856 17569867 22347718 26109357 26109356 24845553 17550304 18057021 20237474 23537049 15632085 18362917 19820115 17510324 25315136 26108605 20966253 20920257 23761445 25244985 24438588 12364791 14747013 17210077 18548007 20566863 26823975
EMBL
BABH01015908
BABH01015909
BABH01015910
AB243445
AB703646
BAO53207.1
+ More
BAO53211.1 KF827820 AHV83756.1 NWSH01002531 PCG68055.1 PCG68056.1 KQ459463 KPJ00566.1 KQ461108 KPJ09099.1 AGBW02009564 OWR50481.1 GEZM01044333 JAV78305.1 CH963847 KRF97725.1 KRF97727.1 KRF97726.1 EDW73275.2 CM002912 KMY98498.1 KMY98495.1 KMY98493.1 AY302598 AE014296 JN836988 AEU17863.1 CH954178 KQS43402.1 EDV50619.2 KQS43403.1 JXUM01037084 KQ561116 KXJ79660.1 CH480815 EDW40778.1 KMY98494.1 JQ015263 AEU17952.1 KMY98497.1 JN400354 AEU04534.1 AGB94296.1 KQS43405.1 KQS43404.1 AGB94297.1 JN814911 AET41695.1 AGB94295.1 CP012525 ALC43651.1 KK853297 KDR08794.1 CH933809 KRG05825.1 CM000159 KRK00950.1 EDW93102.2 KRG05824.1 KRG05826.1 GU566732 ADG84993.1 EDW17891.2 KB631620 ERL84735.1 CH902618 KPU78147.1 EDV39668.2 KPU78144.1 KPU78145.1 KPU78146.1 CH379070 KRT09071.1 OUUW01000002 SPP77351.1 EDY74257.2 KRT09069.1 CH940647 EDW69132.2 CH916366 EDV96219.1 NEVH01005290 PNF38881.1 AJWK01015649 AJWK01015650 GU566733 ADG84994.1 CCAG010013762 CCAG010013763 PNF38879.1 KQ971334 KYB28252.1 ABLF02031858 ABLF02031860 GGMS01004409 MBY73612.1 JXJN01014254 KRT09070.1 KX249692 AOR81469.1 JRES01001077 KNC25718.1 CVRI01000001 CRK86167.1 APCN01005343 AXCM01005377 AXCM01005378 ADMH02000689 ETN65314.1 DS231843 EDS35492.1 AXCN02001921 ATLV01021842 KE525324 KFB46925.1 AAAB01008905 EAA09726.4 EU624401 ACC86138.1 EDS35491.1 AAZO01007075 DS235873 EEB19608.1 APGK01057546 APGK01057547 KB741280 ENN70974.1 GDHC01019225 JAP99403.1
BAO53211.1 KF827820 AHV83756.1 NWSH01002531 PCG68055.1 PCG68056.1 KQ459463 KPJ00566.1 KQ461108 KPJ09099.1 AGBW02009564 OWR50481.1 GEZM01044333 JAV78305.1 CH963847 KRF97725.1 KRF97727.1 KRF97726.1 EDW73275.2 CM002912 KMY98498.1 KMY98495.1 KMY98493.1 AY302598 AE014296 JN836988 AEU17863.1 CH954178 KQS43402.1 EDV50619.2 KQS43403.1 JXUM01037084 KQ561116 KXJ79660.1 CH480815 EDW40778.1 KMY98494.1 JQ015263 AEU17952.1 KMY98497.1 JN400354 AEU04534.1 AGB94296.1 KQS43405.1 KQS43404.1 AGB94297.1 JN814911 AET41695.1 AGB94295.1 CP012525 ALC43651.1 KK853297 KDR08794.1 CH933809 KRG05825.1 CM000159 KRK00950.1 EDW93102.2 KRG05824.1 KRG05826.1 GU566732 ADG84993.1 EDW17891.2 KB631620 ERL84735.1 CH902618 KPU78147.1 EDV39668.2 KPU78144.1 KPU78145.1 KPU78146.1 CH379070 KRT09071.1 OUUW01000002 SPP77351.1 EDY74257.2 KRT09069.1 CH940647 EDW69132.2 CH916366 EDV96219.1 NEVH01005290 PNF38881.1 AJWK01015649 AJWK01015650 GU566733 ADG84994.1 CCAG010013762 CCAG010013763 PNF38879.1 KQ971334 KYB28252.1 ABLF02031858 ABLF02031860 GGMS01004409 MBY73612.1 JXJN01014254 KRT09070.1 KX249692 AOR81469.1 JRES01001077 KNC25718.1 CVRI01000001 CRK86167.1 APCN01005343 AXCM01005377 AXCM01005378 ADMH02000689 ETN65314.1 DS231843 EDS35492.1 AXCN02001921 ATLV01021842 KE525324 KFB46925.1 AAAB01008905 EAA09726.4 EU624401 ACC86138.1 EDS35491.1 AAZO01007075 DS235873 EEB19608.1 APGK01057546 APGK01057547 KB741280 ENN70974.1 GDHC01019225 JAP99403.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000092445
+ More
UP000007798 UP000000803 UP000008711 UP000069940 UP000249989 UP000001292 UP000092553 UP000027135 UP000009192 UP000002282 UP000030742 UP000007801 UP000001819 UP000268350 UP000192221 UP000008792 UP000001070 UP000235965 UP000092461 UP000092444 UP000007266 UP000095300 UP000095301 UP000007819 UP000078200 UP000192223 UP000092443 UP000092460 UP000037069 UP000183832 UP000075920 UP000075840 UP000076407 UP000075885 UP000075882 UP000075884 UP000069272 UP000075883 UP000000673 UP000076408 UP000075900 UP000002320 UP000075886 UP000030765 UP000075902 UP000007062 UP000009046 UP000019118 UP000075881 UP000075880
UP000007798 UP000000803 UP000008711 UP000069940 UP000249989 UP000001292 UP000092553 UP000027135 UP000009192 UP000002282 UP000030742 UP000007801 UP000001819 UP000268350 UP000192221 UP000008792 UP000001070 UP000235965 UP000092461 UP000092444 UP000007266 UP000095300 UP000095301 UP000007819 UP000078200 UP000192223 UP000092443 UP000092460 UP000037069 UP000183832 UP000075920 UP000075840 UP000076407 UP000075885 UP000075882 UP000075884 UP000069272 UP000075883 UP000000673 UP000076408 UP000075900 UP000002320 UP000075886 UP000030765 UP000075902 UP000007062 UP000009046 UP000019118 UP000075881 UP000075880
Pfam
Interpro
IPR020683
Ankyrin_rpt-contain_dom
+ More
IPR036770 Ankyrin_rpt-contain_sf
IPR005821 Ion_trans_dom
IPR002110 Ankyrin_rpt
IPR005178 Ostalpha/TMEM184C
IPR028320 iASPP
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR004838 NHTrfase_class1_PyrdxlP-BS
IPR000796 Asp_trans
IPR004839 Aminotransferase_I/II
IPR015424 PyrdxlP-dep_Trfase
IPR015421 PyrdxlP-dep_Trfase_major
IPR035892 C2_domain_sf
IPR037724 C2E_Ferlin
IPR037721 Ferlin
IPR032362 Ferlin_C
IPR000008 C2_dom
IPR037725 C2F_Ferlin
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR036770 Ankyrin_rpt-contain_sf
IPR005821 Ion_trans_dom
IPR002110 Ankyrin_rpt
IPR005178 Ostalpha/TMEM184C
IPR028320 iASPP
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR004838 NHTrfase_class1_PyrdxlP-BS
IPR000796 Asp_trans
IPR004839 Aminotransferase_I/II
IPR015424 PyrdxlP-dep_Trfase
IPR015421 PyrdxlP-dep_Trfase_major
IPR035892 C2_domain_sf
IPR037724 C2E_Ferlin
IPR037721 Ferlin
IPR032362 Ferlin_C
IPR000008 C2_dom
IPR037725 C2F_Ferlin
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
Gene 3D
ProteinModelPortal
H9IXZ8
W8VUV0
X4YI96
A0A2A4J7J5
A0A2A4J8G5
A0A194QAW8
+ More
A0A194QUV6 A0A212F9S1 A0A1A9ZTS1 A0A1Y1M1U7 A0A0Q9X1C4 A0A0Q9WPV3 A0A0Q9WXY6 B4MMD0 A0A0J9RRT9 A0A0J9UHK2 A0A0J9RRT3 Q7Z020-5 H0UST1 Q7Z020 A0A0Q5UHP3 B3NBM3 A0A0Q5U3H8 A0A182HFJ3 B4HK42 A0A0J9RT54 G9L8C4 A0A0J9RS37 H0USS9 A0A0Q5U9M3 A0A0Q5U5B0 M9PBX8 G8IQ59 A0A0M4EIG3 A0A067QW72 A0A0Q9XMU7 A0A0R1DUU3 B4PFH6 A0A0Q9XBB8 A0A0Q9XL17 B4KZG4 U4U429 A0A0P8XTU6 B3M5X8 A0A0P8Y9L5 A0A0P8XTU9 A0A0P9AJX4 A0A0R3P8W6 A0A3B0JXA1 B5DRW8 A0A0R3P941 A0A1W4V0P1 B4LEP7 B4J3L7 A0A2J7RDI1 A0A1B0CK24 D6NLB6 A0A1B0G3C5 A0A2J7RDG6 A0A139WJZ5 A0A1I8PQV0 A0A1I8PQU6 A0A1I8PQU7 A0A1I8PQP5 A0A1S4FME7 A0A1I8NJU2 J9K0X9 A0A1A9V4K4 A0A2S2Q8S0 A0A1W4WSV1 A0A1A9XBH9 A0A1I8QCJ1 A0A1B0BHB3 A0A0R3P7M5 A0A384TFK4 A0A0L0C0A2 A0A1J1HDZ7 A0A182WML1 A0A182HXS5 A0A182WS30 A0A182PV41 A0A182LHX6 A0A182NNL5 A0A182FED1 A0A182M6W7 W5JRE0 A0A182YBU4 A0A182RQE7 B0W5K9 A0A182Q3M9 A0A084W9N1 A0A182UBZ2 Q7Q966 B3G3I7 B0W5K8 E0W214 N6STG4 A0A146KSD5 A0A182JZJ9 A0A182J3R1
A0A194QUV6 A0A212F9S1 A0A1A9ZTS1 A0A1Y1M1U7 A0A0Q9X1C4 A0A0Q9WPV3 A0A0Q9WXY6 B4MMD0 A0A0J9RRT9 A0A0J9UHK2 A0A0J9RRT3 Q7Z020-5 H0UST1 Q7Z020 A0A0Q5UHP3 B3NBM3 A0A0Q5U3H8 A0A182HFJ3 B4HK42 A0A0J9RT54 G9L8C4 A0A0J9RS37 H0USS9 A0A0Q5U9M3 A0A0Q5U5B0 M9PBX8 G8IQ59 A0A0M4EIG3 A0A067QW72 A0A0Q9XMU7 A0A0R1DUU3 B4PFH6 A0A0Q9XBB8 A0A0Q9XL17 B4KZG4 U4U429 A0A0P8XTU6 B3M5X8 A0A0P8Y9L5 A0A0P8XTU9 A0A0P9AJX4 A0A0R3P8W6 A0A3B0JXA1 B5DRW8 A0A0R3P941 A0A1W4V0P1 B4LEP7 B4J3L7 A0A2J7RDI1 A0A1B0CK24 D6NLB6 A0A1B0G3C5 A0A2J7RDG6 A0A139WJZ5 A0A1I8PQV0 A0A1I8PQU6 A0A1I8PQU7 A0A1I8PQP5 A0A1S4FME7 A0A1I8NJU2 J9K0X9 A0A1A9V4K4 A0A2S2Q8S0 A0A1W4WSV1 A0A1A9XBH9 A0A1I8QCJ1 A0A1B0BHB3 A0A0R3P7M5 A0A384TFK4 A0A0L0C0A2 A0A1J1HDZ7 A0A182WML1 A0A182HXS5 A0A182WS30 A0A182PV41 A0A182LHX6 A0A182NNL5 A0A182FED1 A0A182M6W7 W5JRE0 A0A182YBU4 A0A182RQE7 B0W5K9 A0A182Q3M9 A0A084W9N1 A0A182UBZ2 Q7Q966 B3G3I7 B0W5K8 E0W214 N6STG4 A0A146KSD5 A0A182JZJ9 A0A182J3R1
PDB
5OOY
E-value=5.68698e-09,
Score=138
Ontologies
GO
GO:0005216
GO:0016021
GO:0050960
GO:0043052
GO:0040040
GO:0009416
GO:0006812
GO:0050850
GO:0005261
GO:0097604
GO:0023041
GO:0015276
GO:0046957
GO:0007638
GO:0050965
GO:0034605
GO:0007602
GO:0034703
GO:0010378
GO:0009408
GO:0006816
GO:0005886
GO:0005262
GO:0050968
GO:0001580
GO:0004069
GO:0009058
GO:0006520
GO:0030170
GO:0004252
GO:0005515
GO:0022857
GO:0043565
GO:0015074
GO:0048013
GO:0005003
GO:0005887
GO:0007169
Topology
Subcellular location
Cell membrane
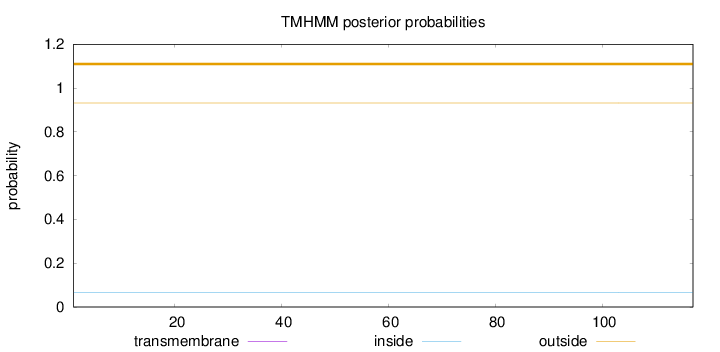
Length:
117
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06783
outside
1 - 117
Population Genetic Test Statistics
Pi
23.416388
Theta
32.191618
Tajima's D
-0.866627
CLR
0.01798
CSRT
0.16249187540623
Interpretation
Uncertain
