Gene
KWMTBOMO00112
Pre Gene Modal
BGIBMGA002127
Annotation
apterous_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 3.442
Sequence
CDS
ATGCTGAAGGAACGCGAGAGCTCAGAAGGATCCCCCGGGGCACCAGACGAGTGCGCAGGCTGCGGAGGCAAAATACAAGATCGATATTACCTCCTAGCTGTGGATCGTCAGTGGCACGGATCCTGCCTGCGCTGCTGCGAATGTCGTCTGCCTTTAGACACCGAACTGACTTGCTTCTCACGGGATGGAAACATATATTGCAAAGAGGATTATTACAGGTTGTTTTGCGTGAAGCGTTGCGGCAGATGCTGTAATGGTATCACTGCCAATGAGCTCGTGATGCGAGCCAGAGACCTGGTCTACCACCTCAACTGCTTCACTTGTGTCGCTTGCGGGACTCAGCTTTCTAAGGGGGATGTGTTCGGAATGAAAGGTGGTCTGGTATACTGTCGTCCTCATTACGACACCGCTTGCTTGGATGACTACTGCGATGAAGAAATGACTAACGGTTATAGGTGTCAAGATATGCACAATGGCGGAAACTCACCCCAGTACTATAGTGGAGGTTCAACTCCAAAAGGTCGGCCGAGGAAGAGAAAGATACCCCACGGCTCTCCGGACGAGCTACAAGCACAAACCATACGGATGGCCAGCTCCGCTCTAGAAATCCTGCATCGTGGCGATCTGTCTTCATCCATGGAGTCTCTGGCTTATGACTCTTCAGTGGCCTCTCCAGGCAGCGTTTCTAGCCACACGCAAAGGACCAAGCGAATGCGAACCAGCTTCAAGCATCACCAGCTACGGACCATGAAATCTTACTTCGCAATCAACCAGAACCCTGATGCGAAAGACTTAAAGCAATTGGCCCAGAAGACTGGACTCTCGAAAAGGGTCTTACAGGTCTGGTTCCAAAATGCGCGCGCAAAATGGCGACGTAACATGATGAGGCAAGAAACGAACGGGGTTAATGGTCATACTTCGATTGTCCCTGGTAACGGTGGAACACCAAGTATAGTTCCCACCGCGCTGATCCTCTCGGAGCCTCTCCAGCCCCTTGAAGATTTGAGGGTACATACTCCCCATCCGATGGCGTTCAATGAACTTTACTGA
Protein
MLKERESSEGSPGAPDECAGCGGKIQDRYYLLAVDRQWHGSCLRCCECRLPLDTELTCFSRDGNIYCKEDYYRLFCVKRCGRCCNGITANELVMRARDLVYHLNCFTCVACGTQLSKGDVFGMKGGLVYCRPHYDTACLDDYCDEEMTNGYRCQDMHNGGNSPQYYSGGSTPKGRPRKRKIPHGSPDELQAQTIRMASSALEILHRGDLSSSMESLAYDSSVASPGSVSSHTQRTKRMRTSFKHHQLRTMKSYFAINQNPDAKDLKQLAQKTGLSKRVLQVWFQNARAKWRRNMMRQETNGVNGHTSIVPGNGGTPSIVPTALILSEPLQPLEDLRVHTPHPMAFNELY
Summary
Uniprot
F2Z7K7
A0A2A4JV72
A0A194QZS9
A0A194Q4V1
A0A212F9S0
A0A139WK10
+ More
C0KZ22 A0A1W4WTN2 A0A154PMF9 E0W1S9 A0A158NLR3 A0A1B6JDG9 A0A088A1G4 A0A0C9RLT4 A0A195FUU8 A0A195CE01 A0A232F860 E2B1F1 K7JAB2 E2C2P6 A0A067R7N9 A0A3L8DK78 A0A026WFM6 A0A3B3D3H9 H2MPF5 A0A3P9LAV2 A0A146RAR2 T1J7L2 A0A2J7QK93 Q4SDH5 Q6JTI8 A0A1S3PI95 A0A060XPK2 A0A1S3NKM7 A0A1L8F6B6 A0A060Y6D2 H2RSS0 A0A1S3PHL4 A0A3B5LKH7 A0A3B5PTQ3 A0A3P9PDA7 A0A3B3QTD5 A0A2I4CRQ8 W5MD70 I3KWI0 A0A147B487 A0A3B4X0S5 A0A3B4U7P8 M3ZNX6 A0A3B4ZKG2 A0A3P8X7Y0 A0A3P8T8B1 A0A1A7XEH9 Q6K1E8 A0A1B8YA12 A0A087XK53 A0A3B4FDN4 A0A3B3XUI4 A0A3B3VIC5 F7E6R5 A0A3P8XBL8 A0A3P8QFZ7 A0A1S3PHH0 A0A3P9CMT3 A0A3B3Z744 B3DGW8 A0A1A8IX63 A0A1A8JUU4 A0A1A8PZ23 A0A1S3AS91 A0A1S3NKL5 A0A151ITR8 A0A3B3TFI4 A0A1A8F3X1 A0A2Y9DJJ7 A0A2U9BR19 O42108 A0A1S3NKM9 G3NH18 H3BHP7 G7NFT9 A0A151MG25 A0A2I0M3N4 A0A1V4J5U9 A0A2I0M3P4 A0A3M0IVB6 A0A2I0U4W5 A0A151MG51 F1M7M2 A0A1A8P2Z1 A0A1A8M022 A0A1A7ZM21 A0A1A8PSG1 A0A1A8BI41 A0A1A8EDL6 A0A1A8V7A0 F6Z9H5 Q543C6
C0KZ22 A0A1W4WTN2 A0A154PMF9 E0W1S9 A0A158NLR3 A0A1B6JDG9 A0A088A1G4 A0A0C9RLT4 A0A195FUU8 A0A195CE01 A0A232F860 E2B1F1 K7JAB2 E2C2P6 A0A067R7N9 A0A3L8DK78 A0A026WFM6 A0A3B3D3H9 H2MPF5 A0A3P9LAV2 A0A146RAR2 T1J7L2 A0A2J7QK93 Q4SDH5 Q6JTI8 A0A1S3PI95 A0A060XPK2 A0A1S3NKM7 A0A1L8F6B6 A0A060Y6D2 H2RSS0 A0A1S3PHL4 A0A3B5LKH7 A0A3B5PTQ3 A0A3P9PDA7 A0A3B3QTD5 A0A2I4CRQ8 W5MD70 I3KWI0 A0A147B487 A0A3B4X0S5 A0A3B4U7P8 M3ZNX6 A0A3B4ZKG2 A0A3P8X7Y0 A0A3P8T8B1 A0A1A7XEH9 Q6K1E8 A0A1B8YA12 A0A087XK53 A0A3B4FDN4 A0A3B3XUI4 A0A3B3VIC5 F7E6R5 A0A3P8XBL8 A0A3P8QFZ7 A0A1S3PHH0 A0A3P9CMT3 A0A3B3Z744 B3DGW8 A0A1A8IX63 A0A1A8JUU4 A0A1A8PZ23 A0A1S3AS91 A0A1S3NKL5 A0A151ITR8 A0A3B3TFI4 A0A1A8F3X1 A0A2Y9DJJ7 A0A2U9BR19 O42108 A0A1S3NKM9 G3NH18 H3BHP7 G7NFT9 A0A151MG25 A0A2I0M3N4 A0A1V4J5U9 A0A2I0M3P4 A0A3M0IVB6 A0A2I0U4W5 A0A151MG51 F1M7M2 A0A1A8P2Z1 A0A1A8M022 A0A1A7ZM21 A0A1A8PSG1 A0A1A8BI41 A0A1A8EDL6 A0A1A8V7A0 F6Z9H5 Q543C6
Pubmed
21296154
26354079
22118469
18362917
19820115
20566863
+ More
21347285 28648823 20798317 20075255 24845553 30249741 24508170 29451363 17554307 15496914 24755649 27762356 21551351 23542700 29240929 25186727 25069045 16470628 20431018 25463417 9299541 15592404 9215903 22002653 22293439 23371554 15057822 19468303 10349636 11042159 11076861 11217851 12466851 16141073
21347285 28648823 20798317 20075255 24845553 30249741 24508170 29451363 17554307 15496914 24755649 27762356 21551351 23542700 29240929 25186727 25069045 16470628 20431018 25463417 9299541 15592404 9215903 22002653 22293439 23371554 15057822 19468303 10349636 11042159 11076861 11217851 12466851 16141073
EMBL
AB543073
BAK19077.1
NWSH01000582
PCG75514.1
KQ461108
KPJ09096.1
+ More
KQ459463 KPJ00563.1 AGBW02009564 OWR50480.1 KQ971334 KYB28253.1 FJ647812 ACN43342.1 KQ434978 KZC13049.1 DS235873 EEB19661.1 ADTU01019691 ADTU01019692 GECU01010365 JAS97341.1 GBYB01014307 JAG84074.1 KQ981219 KYN44425.1 KQ977957 KYM98461.1 NNAY01000712 OXU26845.1 GL444886 EFN60494.1 AAZX01003022 AAZX01003025 AAZX01004767 AAZX01015261 GL452203 EFN77731.1 KK852645 KDR19539.1 QOIP01000007 RLU20746.1 KK107238 EZA54733.1 GCES01120777 JAQ65545.1 JH431936 NEVH01013275 PNF29010.1 CAAE01014638 CAG01307.1 AY307983 AAQ81868.1 FR905368 CDQ78760.1 CM004480 OCT67141.1 FR907771 CDQ87266.1 AHAT01024932 AERX01036361 AERX01036362 AERX01036363 GCES01001151 JAR85172.1 HADW01015101 HADX01006593 SBP16501.1 AY141037 CM004481 AAN41461.1 OCT65600.1 KV460364 OCA19844.1 AYCK01007078 AYCK01007079 AYCK01007080 AAMC01084535 AAMC01084536 AAMC01084537 BC162549 AAI62549.1 HAED01015471 SBR01916.1 HAEE01003652 SBR23672.1 HAEI01003905 SBR86481.1 KQ980982 KYN10789.1 HAEB01006272 HAEC01014675 SBQ52799.1 CP026251 AWP06635.1 AADN05000150 AB005882 BAA21846.1 AFYH01007685 CM001267 EHH23788.1 AKHW03006215 KYO23482.1 AKCR02000043 PKK24291.1 LSYS01008925 OPJ67608.1 PKK24292.1 QRBI01000209 RMB93301.1 KZ506170 PKU41051.1 KYO23483.1 AABR07051808 HAEG01005923 SBR75387.1 HAEF01010445 SBR50395.1 HADY01004876 SBP43361.1 HAEH01008342 SBR84206.1 HADZ01002617 SBP66558.1 HAEA01015533 SBQ44013.1 HAEJ01014990 SBS55447.1 AL929186 BX813320 AK053572 BAC35432.1
KQ459463 KPJ00563.1 AGBW02009564 OWR50480.1 KQ971334 KYB28253.1 FJ647812 ACN43342.1 KQ434978 KZC13049.1 DS235873 EEB19661.1 ADTU01019691 ADTU01019692 GECU01010365 JAS97341.1 GBYB01014307 JAG84074.1 KQ981219 KYN44425.1 KQ977957 KYM98461.1 NNAY01000712 OXU26845.1 GL444886 EFN60494.1 AAZX01003022 AAZX01003025 AAZX01004767 AAZX01015261 GL452203 EFN77731.1 KK852645 KDR19539.1 QOIP01000007 RLU20746.1 KK107238 EZA54733.1 GCES01120777 JAQ65545.1 JH431936 NEVH01013275 PNF29010.1 CAAE01014638 CAG01307.1 AY307983 AAQ81868.1 FR905368 CDQ78760.1 CM004480 OCT67141.1 FR907771 CDQ87266.1 AHAT01024932 AERX01036361 AERX01036362 AERX01036363 GCES01001151 JAR85172.1 HADW01015101 HADX01006593 SBP16501.1 AY141037 CM004481 AAN41461.1 OCT65600.1 KV460364 OCA19844.1 AYCK01007078 AYCK01007079 AYCK01007080 AAMC01084535 AAMC01084536 AAMC01084537 BC162549 AAI62549.1 HAED01015471 SBR01916.1 HAEE01003652 SBR23672.1 HAEI01003905 SBR86481.1 KQ980982 KYN10789.1 HAEB01006272 HAEC01014675 SBQ52799.1 CP026251 AWP06635.1 AADN05000150 AB005882 BAA21846.1 AFYH01007685 CM001267 EHH23788.1 AKHW03006215 KYO23482.1 AKCR02000043 PKK24291.1 LSYS01008925 OPJ67608.1 PKK24292.1 QRBI01000209 RMB93301.1 KZ506170 PKU41051.1 KYO23483.1 AABR07051808 HAEG01005923 SBR75387.1 HAEF01010445 SBR50395.1 HADY01004876 SBP43361.1 HAEH01008342 SBR84206.1 HADZ01002617 SBP66558.1 HAEA01015533 SBQ44013.1 HAEJ01014990 SBS55447.1 AL929186 BX813320 AK053572 BAC35432.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000007266
UP000192223
+ More
UP000076502 UP000009046 UP000005205 UP000005203 UP000078541 UP000078542 UP000215335 UP000000311 UP000002358 UP000008237 UP000027135 UP000279307 UP000053097 UP000261560 UP000001038 UP000265180 UP000265200 UP000235965 UP000007303 UP000087266 UP000193380 UP000186698 UP000005226 UP000261380 UP000002852 UP000242638 UP000261540 UP000192220 UP000018468 UP000005207 UP000261360 UP000261420 UP000261400 UP000265140 UP000265080 UP000008143 UP000028760 UP000261460 UP000261480 UP000261500 UP000265100 UP000265160 UP000261520 UP000079721 UP000078492 UP000248480 UP000246464 UP000000539 UP000007635 UP000008672 UP000050525 UP000053872 UP000190648 UP000269221 UP000002494 UP000000589
UP000076502 UP000009046 UP000005205 UP000005203 UP000078541 UP000078542 UP000215335 UP000000311 UP000002358 UP000008237 UP000027135 UP000279307 UP000053097 UP000261560 UP000001038 UP000265180 UP000265200 UP000235965 UP000007303 UP000087266 UP000193380 UP000186698 UP000005226 UP000261380 UP000002852 UP000242638 UP000261540 UP000192220 UP000018468 UP000005207 UP000261360 UP000261420 UP000261400 UP000265140 UP000265080 UP000008143 UP000028760 UP000261460 UP000261480 UP000261500 UP000265100 UP000265160 UP000261520 UP000079721 UP000078492 UP000248480 UP000246464 UP000000539 UP000007635 UP000008672 UP000050525 UP000053872 UP000190648 UP000269221 UP000002494 UP000000589
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
F2Z7K7
A0A2A4JV72
A0A194QZS9
A0A194Q4V1
A0A212F9S0
A0A139WK10
+ More
C0KZ22 A0A1W4WTN2 A0A154PMF9 E0W1S9 A0A158NLR3 A0A1B6JDG9 A0A088A1G4 A0A0C9RLT4 A0A195FUU8 A0A195CE01 A0A232F860 E2B1F1 K7JAB2 E2C2P6 A0A067R7N9 A0A3L8DK78 A0A026WFM6 A0A3B3D3H9 H2MPF5 A0A3P9LAV2 A0A146RAR2 T1J7L2 A0A2J7QK93 Q4SDH5 Q6JTI8 A0A1S3PI95 A0A060XPK2 A0A1S3NKM7 A0A1L8F6B6 A0A060Y6D2 H2RSS0 A0A1S3PHL4 A0A3B5LKH7 A0A3B5PTQ3 A0A3P9PDA7 A0A3B3QTD5 A0A2I4CRQ8 W5MD70 I3KWI0 A0A147B487 A0A3B4X0S5 A0A3B4U7P8 M3ZNX6 A0A3B4ZKG2 A0A3P8X7Y0 A0A3P8T8B1 A0A1A7XEH9 Q6K1E8 A0A1B8YA12 A0A087XK53 A0A3B4FDN4 A0A3B3XUI4 A0A3B3VIC5 F7E6R5 A0A3P8XBL8 A0A3P8QFZ7 A0A1S3PHH0 A0A3P9CMT3 A0A3B3Z744 B3DGW8 A0A1A8IX63 A0A1A8JUU4 A0A1A8PZ23 A0A1S3AS91 A0A1S3NKL5 A0A151ITR8 A0A3B3TFI4 A0A1A8F3X1 A0A2Y9DJJ7 A0A2U9BR19 O42108 A0A1S3NKM9 G3NH18 H3BHP7 G7NFT9 A0A151MG25 A0A2I0M3N4 A0A1V4J5U9 A0A2I0M3P4 A0A3M0IVB6 A0A2I0U4W5 A0A151MG51 F1M7M2 A0A1A8P2Z1 A0A1A8M022 A0A1A7ZM21 A0A1A8PSG1 A0A1A8BI41 A0A1A8EDL6 A0A1A8V7A0 F6Z9H5 Q543C6
C0KZ22 A0A1W4WTN2 A0A154PMF9 E0W1S9 A0A158NLR3 A0A1B6JDG9 A0A088A1G4 A0A0C9RLT4 A0A195FUU8 A0A195CE01 A0A232F860 E2B1F1 K7JAB2 E2C2P6 A0A067R7N9 A0A3L8DK78 A0A026WFM6 A0A3B3D3H9 H2MPF5 A0A3P9LAV2 A0A146RAR2 T1J7L2 A0A2J7QK93 Q4SDH5 Q6JTI8 A0A1S3PI95 A0A060XPK2 A0A1S3NKM7 A0A1L8F6B6 A0A060Y6D2 H2RSS0 A0A1S3PHL4 A0A3B5LKH7 A0A3B5PTQ3 A0A3P9PDA7 A0A3B3QTD5 A0A2I4CRQ8 W5MD70 I3KWI0 A0A147B487 A0A3B4X0S5 A0A3B4U7P8 M3ZNX6 A0A3B4ZKG2 A0A3P8X7Y0 A0A3P8T8B1 A0A1A7XEH9 Q6K1E8 A0A1B8YA12 A0A087XK53 A0A3B4FDN4 A0A3B3XUI4 A0A3B3VIC5 F7E6R5 A0A3P8XBL8 A0A3P8QFZ7 A0A1S3PHH0 A0A3P9CMT3 A0A3B3Z744 B3DGW8 A0A1A8IX63 A0A1A8JUU4 A0A1A8PZ23 A0A1S3AS91 A0A1S3NKL5 A0A151ITR8 A0A3B3TFI4 A0A1A8F3X1 A0A2Y9DJJ7 A0A2U9BR19 O42108 A0A1S3NKM9 G3NH18 H3BHP7 G7NFT9 A0A151MG25 A0A2I0M3N4 A0A1V4J5U9 A0A2I0M3P4 A0A3M0IVB6 A0A2I0U4W5 A0A151MG51 F1M7M2 A0A1A8P2Z1 A0A1A8M022 A0A1A7ZM21 A0A1A8PSG1 A0A1A8BI41 A0A1A8EDL6 A0A1A8V7A0 F6Z9H5 Q543C6
PDB
2DMQ
E-value=7.87841e-31,
Score=333
Ontologies
GO
GO:0006355
GO:0043565
GO:0046872
GO:0005634
GO:0000981
GO:0007409
GO:0048665
GO:0016740
GO:0003677
GO:0021794
GO:0021554
GO:0021537
GO:0031290
GO:0071632
GO:0043010
GO:0030182
GO:0000977
GO:0045944
GO:0007411
GO:0007634
GO:0010468
GO:0009416
GO:0030900
GO:0007420
GO:0008045
GO:0001228
GO:0000978
GO:0021978
GO:0021987
GO:2000678
GO:0048675
GO:0001843
GO:0009953
GO:0021522
GO:0060041
GO:0021772
GO:0003682
GO:0045199
GO:0045814
GO:0007498
GO:0050768
GO:0008134
GO:2000179
GO:0001942
GO:0008270
GO:0015074
GO:0048013
GO:0005003
GO:0005887
GO:0007169
GO:0016021
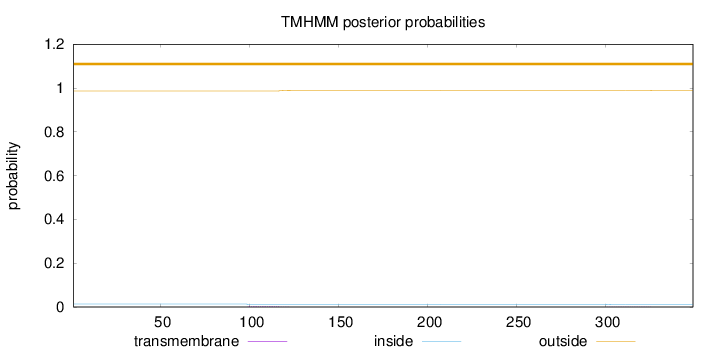
Topology
Subcellular location
Nucleus
Length:
349
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06563
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01401
outside
1 - 349
Population Genetic Test Statistics
Pi
18.056493
Theta
21.72665
Tajima's D
-1.316005
CLR
0.008407
CSRT
0.0853957302134893
Interpretation
Uncertain
