Gene
KWMTBOMO00103
Pre Gene Modal
BGIBMGA002015
Annotation
PREDICTED:_succinate_dehydrogenase_assembly_factor_3?_mitochondrial_[Plutella_xylostella]
Full name
Succinate dehydrogenase assembly factor 3, mitochondrial
+ More
Cysteine dioxygenase
Cysteine dioxygenase
Location in the cell
Cytoplasmic Reliability : 1.347 Mitochondrial Reliability : 1.969
Sequence
CDS
ATGTCATCACCCGAACATGTGTCGCGGGTTCGCCGGCTCTACAAATTGATCTTCCGGGTTCACCGGGGCTTGCCGCCCGAACTCCGTGTCCTGGGAGACAACTACGCCAGGGACGAGTTCAAGCGACACAAAAAATGTAATAAGGACGAGGCTAAGATTTTTCTCAACGAATGGACGGACTACGCCATTAACCTTGCCAAGCAGATGAAGCCGCTGCACCAAGCCAAGAACAAAATGGTCGGCGGCGTCATCGGAGCCTGA
Protein
MSSPEHVSRVRRLYKLIFRVHRGLPPELRVLGDNYARDEFKRHKKCNKDEAKIFLNEWTDYAINLAKQMKPLHQAKNKMVGGVIGA
Summary
Description
Plays an essential role in the assembly of succinate dehydrogenase (SDH), an enzyme complex (also referred to as respiratory complex II) that is a component of both the tricarboxylic acid (TCA) cycle and the mitochondrial electron transport chain, and which couples the oxidation of succinate to fumarate with the reduction of ubiquinone (coenzyme Q) to ubiquinol. Promotes maturation of the iron-sulfur protein subunit SdhB of the SDH catalytic dimer, protecting it from the deleterious effects of oxidants.
Catalytic Activity
L-cysteine + O2 = 3-sulfino-L-alanine + H(+)
Cofactor
Fe cation
Subunit
Interacts with SdhB within an SdhA-SdhB subcomplex.
Similarity
Belongs to the complex I LYR family. SDHAF3 subfamily.
Belongs to the cysteine dioxygenase family.
Belongs to the cysteine dioxygenase family.
Keywords
Chaperone
Complete proteome
Mitochondrion
Reference proteome
Transit peptide
Feature
chain Succinate dehydrogenase assembly factor 3, mitochondrial
Uniprot
H9IXN2
A0A2H1VYT1
A0A2W1C0M8
A0A212F9R6
I4DK21
A0A194QVU9
+ More
A0A194Q4T8 A0A1E1WPD3 A0A232F4Q7 K7IV52 A0A2A3E897 A0A0L7LP77 E2B2T3 A0A026WMZ5 E2A5K4 A0A3L8DES3 A0A1S4FG83 A0A195DEM4 A0A1S3DTU4 B0X1Y1 A0A151WY44 Q172I4 A0A195FNK9 A0A1Q3F9M1 A0A182HEU2 A0A0J7NHD2 A0A195CVS9 A0A1B6CLA6 R7UN63 A0A2J7RQ91 A0A0C9QX20 A0A139WFT2 B4M4D3 B4K680 A0A0N0U3A2 A0A067R8H3 B4QX20 A0A1B2AJN1 Q8SZ16 B4PQL1 A0A2S2PB52 K1PRJ9 B3M2T3 A0A336M5K0 B3P135 B4IBU6 A0A1W4UKP6 A0A1B6EK63 B4JFC5 A0A0K2TQI0 B4N664 A0A1Y1NAM9 E0VAE2 A0A3B0JN23 E9H6U3 A0A2S2Q373 A0A1B6J7Q3 A0A1J1IH16 C4WV75 A0A0P5X7N6 A0A164GA42 A0A0P5NY13 A0A1B6MU64 B5DVG5 A0A1B6JZR4 A0A267EJ60 A0A182T500 A0A182YN10 A0A0P4YNJ0 A0A0P5IHW7 A0A0P5B9K6 W8CDR3 A0A0P5QPZ4 A0A1S3H7W1 A0A1I8NI70 A0A0P5DNA4 C1BQS9 B4GFC2 A0A0P4X2S0 A0A034WU70 A0A0C9R2I2 A0A182JXD8 A0A0K8WEG3 A0A0N8ELD8 A0A0L7R6K7 A0A1I8QFE9 A0A0R3STB6 A0A2Y9D194 A0A182XJE0 A0NG50 A0A182I0W5 A0A0A9YDQ3 A0A182VSS5 A0A182RAE9 A0A182MK21 A0A1B0GDG4 A0A1A9UIK1 A0A1A9ZHV6 A0A0B7A3T3 A0A1W4X2L2
A0A194Q4T8 A0A1E1WPD3 A0A232F4Q7 K7IV52 A0A2A3E897 A0A0L7LP77 E2B2T3 A0A026WMZ5 E2A5K4 A0A3L8DES3 A0A1S4FG83 A0A195DEM4 A0A1S3DTU4 B0X1Y1 A0A151WY44 Q172I4 A0A195FNK9 A0A1Q3F9M1 A0A182HEU2 A0A0J7NHD2 A0A195CVS9 A0A1B6CLA6 R7UN63 A0A2J7RQ91 A0A0C9QX20 A0A139WFT2 B4M4D3 B4K680 A0A0N0U3A2 A0A067R8H3 B4QX20 A0A1B2AJN1 Q8SZ16 B4PQL1 A0A2S2PB52 K1PRJ9 B3M2T3 A0A336M5K0 B3P135 B4IBU6 A0A1W4UKP6 A0A1B6EK63 B4JFC5 A0A0K2TQI0 B4N664 A0A1Y1NAM9 E0VAE2 A0A3B0JN23 E9H6U3 A0A2S2Q373 A0A1B6J7Q3 A0A1J1IH16 C4WV75 A0A0P5X7N6 A0A164GA42 A0A0P5NY13 A0A1B6MU64 B5DVG5 A0A1B6JZR4 A0A267EJ60 A0A182T500 A0A182YN10 A0A0P4YNJ0 A0A0P5IHW7 A0A0P5B9K6 W8CDR3 A0A0P5QPZ4 A0A1S3H7W1 A0A1I8NI70 A0A0P5DNA4 C1BQS9 B4GFC2 A0A0P4X2S0 A0A034WU70 A0A0C9R2I2 A0A182JXD8 A0A0K8WEG3 A0A0N8ELD8 A0A0L7R6K7 A0A1I8QFE9 A0A0R3STB6 A0A2Y9D194 A0A182XJE0 A0NG50 A0A182I0W5 A0A0A9YDQ3 A0A182VSS5 A0A182RAE9 A0A182MK21 A0A1B0GDG4 A0A1A9UIK1 A0A1A9ZHV6 A0A0B7A3T3 A0A1W4X2L2
EC Number
1.13.11.20
Pubmed
19121390
28756777
22118469
22651552
26354079
28648823
+ More
20075255 26227816 20798317 24508170 30249741 17510324 26483478 23254933 18362917 19820115 17994087 24845553 10731132 12537572 12537569 24954417 17550304 22992520 28004739 20566863 21292972 15632085 25244985 24495485 25315136 25348373 20966253 12364791 14747013 17210077 25401762
20075255 26227816 20798317 24508170 30249741 17510324 26483478 23254933 18362917 19820115 17994087 24845553 10731132 12537572 12537569 24954417 17550304 22992520 28004739 20566863 21292972 15632085 25244985 24495485 25315136 25348373 20966253 12364791 14747013 17210077 25401762
EMBL
BABH01015863
ODYU01005124
SOQ45682.1
KZ149901
PZC78516.1
AGBW02009564
+ More
OWR50474.1 AK401639 BAM18261.1 KQ461108 KPJ09090.1 KQ459463 KPJ00557.1 GDQN01002141 JAT88913.1 NNAY01000950 OXU25761.1 AAZX01007538 KZ288326 PBC27977.1 JTDY01000423 KOB77257.1 GL445235 EFN89998.1 KK107148 EZA57442.1 GL436993 EFN71287.1 QOIP01000009 RLU18389.1 KQ980934 KYN11326.1 DS232274 EDS38911.1 KQ982652 KYQ52808.1 CH477436 EAT40942.1 KQ981382 KYN42140.1 GFDL01010832 JAV24213.1 JXUM01036159 KQ561085 KXJ79760.1 LBMM01005007 KMQ91920.1 KQ977279 KYN04264.1 GEDC01023068 GEDC01008935 GEDC01005562 JAS14230.1 JAS28363.1 JAS31736.1 AMQN01006894 KB299468 ELU07974.1 NEVH01001345 PNF42998.1 GBYB01000235 JAG70002.1 KQ971351 KYB26786.1 CH940652 EDW59494.1 CH933806 EDW14130.1 KQ435896 KOX69328.1 KK852811 KDR15889.1 CM000364 EDX12674.1 KX531340 ANY27150.1 AE014297 AY071188 CM000160 EDW97307.1 GGMR01014064 MBY26683.1 JH816424 EKC21454.1 CH902617 EDV43463.1 UFQT01000186 SSX21308.1 CH954181 EDV49154.1 CH480827 EDW44854.1 GECZ01031466 JAS38303.1 CH916369 EDV93406.1 HACA01010704 CDW28065.1 CH964154 EDW79853.1 GEZM01013315 GEZM01013313 JAV92667.1 DS235005 EEB10348.1 OUUW01000007 SPP83624.1 GL732598 EFX72572.1 GGMS01002933 MBY72136.1 GECU01026791 GECU01012518 JAS80915.1 JAS95188.1 CVRI01000048 CRK98836.1 ABLF02037413 AK341389 BAH71795.1 GDIP01076217 JAM27498.1 LRGB01016081 KZR98715.1 GDIQ01146148 JAL05578.1 GEBQ01000520 JAT39457.1 CM000070 EDY68311.1 GECU01003333 JAT04374.1 NIVC01002109 PAA60812.1 GDIP01226252 JAI97149.1 GDIQ01216176 JAK35549.1 GDIP01188354 JAJ35048.1 GAMC01001498 GAMC01001496 JAC05060.1 GDIQ01114008 JAL37718.1 GDIP01153598 JAJ69804.1 BT076958 ACO11382.1 CH479182 EDW34307.1 GDIP01247224 JAI76177.1 GAKP01001237 JAC57715.1 GBYB01007052 JAG76819.1 GDHF01002751 JAI49563.1 GDIQ01015364 JAN79373.1 KQ414646 KOC66510.1 UYSG01011113 VDL60948.1 AAAB01008980 EAU76145.2 APCN01005279 GBHO01012402 GBRD01010890 JAG31202.1 JAG54934.1 AXCM01000163 CCAG010011880 HACG01027851 CEK74716.1
OWR50474.1 AK401639 BAM18261.1 KQ461108 KPJ09090.1 KQ459463 KPJ00557.1 GDQN01002141 JAT88913.1 NNAY01000950 OXU25761.1 AAZX01007538 KZ288326 PBC27977.1 JTDY01000423 KOB77257.1 GL445235 EFN89998.1 KK107148 EZA57442.1 GL436993 EFN71287.1 QOIP01000009 RLU18389.1 KQ980934 KYN11326.1 DS232274 EDS38911.1 KQ982652 KYQ52808.1 CH477436 EAT40942.1 KQ981382 KYN42140.1 GFDL01010832 JAV24213.1 JXUM01036159 KQ561085 KXJ79760.1 LBMM01005007 KMQ91920.1 KQ977279 KYN04264.1 GEDC01023068 GEDC01008935 GEDC01005562 JAS14230.1 JAS28363.1 JAS31736.1 AMQN01006894 KB299468 ELU07974.1 NEVH01001345 PNF42998.1 GBYB01000235 JAG70002.1 KQ971351 KYB26786.1 CH940652 EDW59494.1 CH933806 EDW14130.1 KQ435896 KOX69328.1 KK852811 KDR15889.1 CM000364 EDX12674.1 KX531340 ANY27150.1 AE014297 AY071188 CM000160 EDW97307.1 GGMR01014064 MBY26683.1 JH816424 EKC21454.1 CH902617 EDV43463.1 UFQT01000186 SSX21308.1 CH954181 EDV49154.1 CH480827 EDW44854.1 GECZ01031466 JAS38303.1 CH916369 EDV93406.1 HACA01010704 CDW28065.1 CH964154 EDW79853.1 GEZM01013315 GEZM01013313 JAV92667.1 DS235005 EEB10348.1 OUUW01000007 SPP83624.1 GL732598 EFX72572.1 GGMS01002933 MBY72136.1 GECU01026791 GECU01012518 JAS80915.1 JAS95188.1 CVRI01000048 CRK98836.1 ABLF02037413 AK341389 BAH71795.1 GDIP01076217 JAM27498.1 LRGB01016081 KZR98715.1 GDIQ01146148 JAL05578.1 GEBQ01000520 JAT39457.1 CM000070 EDY68311.1 GECU01003333 JAT04374.1 NIVC01002109 PAA60812.1 GDIP01226252 JAI97149.1 GDIQ01216176 JAK35549.1 GDIP01188354 JAJ35048.1 GAMC01001498 GAMC01001496 JAC05060.1 GDIQ01114008 JAL37718.1 GDIP01153598 JAJ69804.1 BT076958 ACO11382.1 CH479182 EDW34307.1 GDIP01247224 JAI76177.1 GAKP01001237 JAC57715.1 GBYB01007052 JAG76819.1 GDHF01002751 JAI49563.1 GDIQ01015364 JAN79373.1 KQ414646 KOC66510.1 UYSG01011113 VDL60948.1 AAAB01008980 EAU76145.2 APCN01005279 GBHO01012402 GBRD01010890 JAG31202.1 JAG54934.1 AXCM01000163 CCAG010011880 HACG01027851 CEK74716.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000215335
UP000002358
+ More
UP000242457 UP000037510 UP000008237 UP000053097 UP000000311 UP000279307 UP000078492 UP000079169 UP000002320 UP000075809 UP000008820 UP000078541 UP000069940 UP000249989 UP000036403 UP000078542 UP000014760 UP000235965 UP000007266 UP000008792 UP000009192 UP000053105 UP000027135 UP000000304 UP000000803 UP000002282 UP000005408 UP000007801 UP000008711 UP000001292 UP000192221 UP000001070 UP000007798 UP000009046 UP000268350 UP000000305 UP000183832 UP000007819 UP000076858 UP000001819 UP000215902 UP000075901 UP000076408 UP000085678 UP000095301 UP000008744 UP000075881 UP000053825 UP000095300 UP000046397 UP000274504 UP000075882 UP000076407 UP000007062 UP000075840 UP000075920 UP000075900 UP000075883 UP000092444 UP000078200 UP000092445 UP000192223
UP000242457 UP000037510 UP000008237 UP000053097 UP000000311 UP000279307 UP000078492 UP000079169 UP000002320 UP000075809 UP000008820 UP000078541 UP000069940 UP000249989 UP000036403 UP000078542 UP000014760 UP000235965 UP000007266 UP000008792 UP000009192 UP000053105 UP000027135 UP000000304 UP000000803 UP000002282 UP000005408 UP000007801 UP000008711 UP000001292 UP000192221 UP000001070 UP000007798 UP000009046 UP000268350 UP000000305 UP000183832 UP000007819 UP000076858 UP000001819 UP000215902 UP000075901 UP000076408 UP000085678 UP000095301 UP000008744 UP000075881 UP000053825 UP000095300 UP000046397 UP000274504 UP000075882 UP000076407 UP000007062 UP000075840 UP000075920 UP000075900 UP000075883 UP000092444 UP000078200 UP000092445 UP000192223
Interpro
Gene 3D
ProteinModelPortal
H9IXN2
A0A2H1VYT1
A0A2W1C0M8
A0A212F9R6
I4DK21
A0A194QVU9
+ More
A0A194Q4T8 A0A1E1WPD3 A0A232F4Q7 K7IV52 A0A2A3E897 A0A0L7LP77 E2B2T3 A0A026WMZ5 E2A5K4 A0A3L8DES3 A0A1S4FG83 A0A195DEM4 A0A1S3DTU4 B0X1Y1 A0A151WY44 Q172I4 A0A195FNK9 A0A1Q3F9M1 A0A182HEU2 A0A0J7NHD2 A0A195CVS9 A0A1B6CLA6 R7UN63 A0A2J7RQ91 A0A0C9QX20 A0A139WFT2 B4M4D3 B4K680 A0A0N0U3A2 A0A067R8H3 B4QX20 A0A1B2AJN1 Q8SZ16 B4PQL1 A0A2S2PB52 K1PRJ9 B3M2T3 A0A336M5K0 B3P135 B4IBU6 A0A1W4UKP6 A0A1B6EK63 B4JFC5 A0A0K2TQI0 B4N664 A0A1Y1NAM9 E0VAE2 A0A3B0JN23 E9H6U3 A0A2S2Q373 A0A1B6J7Q3 A0A1J1IH16 C4WV75 A0A0P5X7N6 A0A164GA42 A0A0P5NY13 A0A1B6MU64 B5DVG5 A0A1B6JZR4 A0A267EJ60 A0A182T500 A0A182YN10 A0A0P4YNJ0 A0A0P5IHW7 A0A0P5B9K6 W8CDR3 A0A0P5QPZ4 A0A1S3H7W1 A0A1I8NI70 A0A0P5DNA4 C1BQS9 B4GFC2 A0A0P4X2S0 A0A034WU70 A0A0C9R2I2 A0A182JXD8 A0A0K8WEG3 A0A0N8ELD8 A0A0L7R6K7 A0A1I8QFE9 A0A0R3STB6 A0A2Y9D194 A0A182XJE0 A0NG50 A0A182I0W5 A0A0A9YDQ3 A0A182VSS5 A0A182RAE9 A0A182MK21 A0A1B0GDG4 A0A1A9UIK1 A0A1A9ZHV6 A0A0B7A3T3 A0A1W4X2L2
A0A194Q4T8 A0A1E1WPD3 A0A232F4Q7 K7IV52 A0A2A3E897 A0A0L7LP77 E2B2T3 A0A026WMZ5 E2A5K4 A0A3L8DES3 A0A1S4FG83 A0A195DEM4 A0A1S3DTU4 B0X1Y1 A0A151WY44 Q172I4 A0A195FNK9 A0A1Q3F9M1 A0A182HEU2 A0A0J7NHD2 A0A195CVS9 A0A1B6CLA6 R7UN63 A0A2J7RQ91 A0A0C9QX20 A0A139WFT2 B4M4D3 B4K680 A0A0N0U3A2 A0A067R8H3 B4QX20 A0A1B2AJN1 Q8SZ16 B4PQL1 A0A2S2PB52 K1PRJ9 B3M2T3 A0A336M5K0 B3P135 B4IBU6 A0A1W4UKP6 A0A1B6EK63 B4JFC5 A0A0K2TQI0 B4N664 A0A1Y1NAM9 E0VAE2 A0A3B0JN23 E9H6U3 A0A2S2Q373 A0A1B6J7Q3 A0A1J1IH16 C4WV75 A0A0P5X7N6 A0A164GA42 A0A0P5NY13 A0A1B6MU64 B5DVG5 A0A1B6JZR4 A0A267EJ60 A0A182T500 A0A182YN10 A0A0P4YNJ0 A0A0P5IHW7 A0A0P5B9K6 W8CDR3 A0A0P5QPZ4 A0A1S3H7W1 A0A1I8NI70 A0A0P5DNA4 C1BQS9 B4GFC2 A0A0P4X2S0 A0A034WU70 A0A0C9R2I2 A0A182JXD8 A0A0K8WEG3 A0A0N8ELD8 A0A0L7R6K7 A0A1I8QFE9 A0A0R3STB6 A0A2Y9D194 A0A182XJE0 A0NG50 A0A182I0W5 A0A0A9YDQ3 A0A182VSS5 A0A182RAE9 A0A182MK21 A0A1B0GDG4 A0A1A9UIK1 A0A1A9ZHV6 A0A0B7A3T3 A0A1W4X2L2
Ontologies
GO
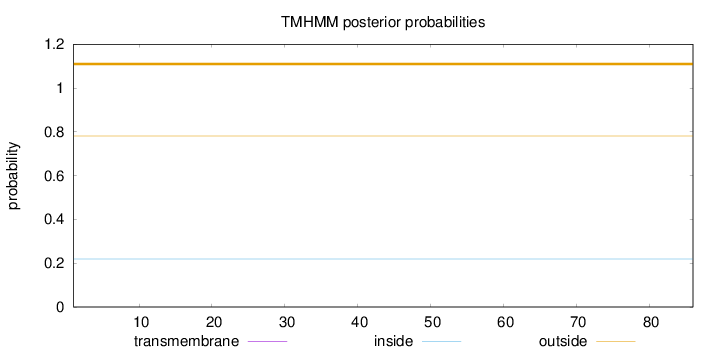
Topology
Subcellular location
Mitochondrion matrix
Length:
86
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0002
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.21942
outside
1 - 86
Population Genetic Test Statistics
Pi
2.258291
Theta
3.909269
Tajima's D
-2.083699
CLR
0.043009
CSRT
0.00649967501624919
Interpretation
Possibly Positive selection
