Gene
KWMTBOMO00097
Pre Gene Modal
BGIBMGA002115
Annotation
PREDICTED:_uncharacterized_protein_CG1161_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 2.204
Sequence
CDS
ATGTATCTGAAATATATTATTTTAGTGTTTCTTGGAACGTGCATCTTCTTTGTGTCTGGCACTTTTGAGAACAAACGTTGCAAGTGTGTGTGTCCGAGCCCCGCAGCTGTCTTCAATAACACTGCTGACACAGGACGCAGTCCGTTTATCGACAATGTGCCTCCTAATAAATGCAACTGTGAAGGCCTAGTGCTACCCCGAATTGGAGACCAACTCAAAGATCGTGCTCAAGAATTTTGTCCCCGTTGTCAGTGCAAATATGAGAACAGAAACATCACAATTATAAAGGTAGTGGTGATTATTGTTATCTGGGTGATCATGTTGCTGGTGGGGTACATGGGTTTCCTGATTTGTTTGGATCCTCTAATCAACAAGCGAGCCATGGCATCATATCAGGAGCATACCAATGAGGATGATGACAACCCTATACCAGGAACGTCTCAGACGATGGGTGCAACTCCCCGCGGGAATGTTTTGAACCGGGTTACCCATCAGCAGGACAAGTGGAAGCGTCAGGTCCGTGAGCAGCGTCGCAACATTTATGATAGACACACTATGTTGAACTAG
Protein
MYLKYIILVFLGTCIFFVSGTFENKRCKCVCPSPAAVFNNTADTGRSPFIDNVPPNKCNCEGLVLPRIGDQLKDRAQEFCPRCQCKYENRNITIIKVVVIIVIWVIMLLVGYMGFLICLDPLINKRAMASYQEHTNEDDDNPIPGTSQTMGATPRGNVLNRVTHQQDKWKRQVREQRRNIYDRHTMLN
Summary
Uniprot
H9IXY2
A0A2W1BHP3
A0A212F9N5
A0A194Q5B8
A0A2A4JLL6
A0A194QUE4
+ More
B0W3V1 A0A1Q3FIC0 Q16ZG7 A0A1L8D9K5 R4V1N3 A0A023EI23 A0A1B0C917 A0A182JTP2 A0A182G242 A0A084VQU8 T1D4Y0 A0A067RCE1 A0A182QGI4 A0A182P232 A0A195FGP2 A0A182U862 Q7QJ43 V5GYC8 F4X7B8 A0A182I2U2 E0V9V8 A0A151J637 A0A0L7QY08 A0A026WGP3 A0A195BM69 A0A158ND58 A0A2M4C0H1 A0A2M3Z3I6 U5EEX9 D6WIJ0 A0A2M4AIW0 E2AAQ3 B6DE27 A0A088A9P6 E2BRR8 A0A0N0BCR8 A0A0C9QIM6 K7J6B4 A0A151IFD4 A0A1Y1MJD3 A0A182T1X6 A0A1Y1MM90 A0A2A3EP73 A0A232F277 A0A182M0Y9 A0A1E1W4S2 A0A0P5LDQ5 A0A0P5JRT6 A0A087UBC8 A0A0P4XYL9 E9GXY2 A0A0P5RWN6 N6UDP3 A0A0P5B5C8 A0A0P5ZIB1 A0A0P4ZAW5 A0A0P5TTI8 A0A0P5L2P6 A0A1B6HWZ5 A0A1B6ERE5 A0A1J1IIZ7 A0A0U1U058 A0A131Y6C1 A0A1Z5KUB7 T1J2U0 A0A0K8R3I8 L7LYE8 A0A2J7RK74 A0A293MNI7 V5H0A1 A0A131Z036 A0A224Z6L4 B7PD07 A0A293LHM2 A0A0P5SD31 A0A0P6HZJ0 A0A023FUE0 A0A1B0DJG7 A0A023GPA1 A0A1E1WZ69 A0A0C9R669 A0A023FMQ1 A0A1E1XUN0 A0A023FL44 A0A1B6BY24 A0A226E5T3 A0A1Y1MJ29 A0A336ML15 A0A336L3M2 A0A0P5S8I4 A0A0A9YFX8 A0A0P5BP59 A0A1W7R544
B0W3V1 A0A1Q3FIC0 Q16ZG7 A0A1L8D9K5 R4V1N3 A0A023EI23 A0A1B0C917 A0A182JTP2 A0A182G242 A0A084VQU8 T1D4Y0 A0A067RCE1 A0A182QGI4 A0A182P232 A0A195FGP2 A0A182U862 Q7QJ43 V5GYC8 F4X7B8 A0A182I2U2 E0V9V8 A0A151J637 A0A0L7QY08 A0A026WGP3 A0A195BM69 A0A158ND58 A0A2M4C0H1 A0A2M3Z3I6 U5EEX9 D6WIJ0 A0A2M4AIW0 E2AAQ3 B6DE27 A0A088A9P6 E2BRR8 A0A0N0BCR8 A0A0C9QIM6 K7J6B4 A0A151IFD4 A0A1Y1MJD3 A0A182T1X6 A0A1Y1MM90 A0A2A3EP73 A0A232F277 A0A182M0Y9 A0A1E1W4S2 A0A0P5LDQ5 A0A0P5JRT6 A0A087UBC8 A0A0P4XYL9 E9GXY2 A0A0P5RWN6 N6UDP3 A0A0P5B5C8 A0A0P5ZIB1 A0A0P4ZAW5 A0A0P5TTI8 A0A0P5L2P6 A0A1B6HWZ5 A0A1B6ERE5 A0A1J1IIZ7 A0A0U1U058 A0A131Y6C1 A0A1Z5KUB7 T1J2U0 A0A0K8R3I8 L7LYE8 A0A2J7RK74 A0A293MNI7 V5H0A1 A0A131Z036 A0A224Z6L4 B7PD07 A0A293LHM2 A0A0P5SD31 A0A0P6HZJ0 A0A023FUE0 A0A1B0DJG7 A0A023GPA1 A0A1E1WZ69 A0A0C9R669 A0A023FMQ1 A0A1E1XUN0 A0A023FL44 A0A1B6BY24 A0A226E5T3 A0A1Y1MJ29 A0A336ML15 A0A336L3M2 A0A0P5S8I4 A0A0A9YFX8 A0A0P5BP59 A0A1W7R544
Pubmed
19121390
28756777
22118469
26354079
17510324
24945155
+ More
26483478 24438588 24330624 24845553 12364791 14747013 17210077 21719571 20566863 24508170 30249741 21347285 18362917 19820115 20798317 19178717 20920257 23761445 20075255 28004739 28648823 21292972 23537049 28528879 25576852 25765539 26830274 28797301 28503490 26131772 29209593 25401762 26823975
26483478 24438588 24330624 24845553 12364791 14747013 17210077 21719571 20566863 24508170 30249741 21347285 18362917 19820115 20798317 19178717 20920257 23761445 20075255 28004739 28648823 21292972 23537049 28528879 25576852 25765539 26830274 28797301 28503490 26131772 29209593 25401762 26823975
EMBL
BABH01015823
KZ150037
PZC74619.1
AGBW02009564
OWR50419.1
KQ459463
+ More
KPJ00554.1 NWSH01001084 PCG72709.1 KQ461108 KPJ09087.1 DS231833 EDS32248.1 GFDL01007751 JAV27294.1 CH477491 EAT40023.1 GFDF01011074 JAV03010.1 KC740768 AGM32592.1 GAPW01005057 JAC08541.1 AJWK01001776 JXUM01039167 JXUM01039168 KQ561193 KXJ79409.1 ATLV01015326 KE525006 KFB40342.1 GALA01000788 JAA94064.1 KK852598 KDR20537.1 AXCN02000544 KQ981560 KYN39850.1 AAAB01008807 EAA04310.4 GALX01001614 JAB66852.1 GL888828 EGI57730.1 APCN01000180 DS234996 EEB10164.1 KQ979878 KYN18691.1 KQ414699 KOC63429.1 KK107260 QOIP01000001 EZA54249.1 RLU26241.1 KQ976438 KYM86874.1 ADTU01012403 GGFJ01009623 MBW58764.1 GGFM01002314 MBW23065.1 GANO01004120 JAB55751.1 KQ971334 EEZ99657.2 GGFK01007392 MBW40713.1 GL438137 EFN69503.1 EU934396 ADMH02002078 ACI30174.1 ETN59499.1 GL449994 EFN81610.1 KQ435891 KOX69404.1 GBYB01000472 JAG70239.1 KQ977796 KYM99618.1 GEZM01030096 JAV85811.1 GEZM01030097 JAV85810.1 KZ288212 PBC32831.1 NNAY01001263 OXU24568.1 AXCM01004785 GDQN01009115 JAT81939.1 GDIQ01178681 JAK73044.1 GDIQ01199578 JAK52147.1 KK119091 KFM74667.1 GDIP01235055 JAI88346.1 GL732574 EFX75498.1 GDIQ01095263 JAL56463.1 APGK01026508 KB740635 KB631992 ENN79825.1 ERL87768.1 GDIP01189400 JAJ34002.1 GDIP01043165 LRGB01000872 JAM60550.1 KZS15356.1 GDIP01215855 JAJ07547.1 GDIP01121757 JAL81957.1 GDIQ01200876 JAK50849.1 GECU01028502 JAS79204.1 GECZ01029289 GECZ01027354 JAS40480.1 JAS42415.1 CVRI01000054 CRL00215.1 EU252459 ACD12008.1 GEFM01000782 JAP75014.1 GFJQ02008283 JAV98686.1 JH431809 GADI01008360 JAA65448.1 GACK01008412 JAA56622.1 NEVH01002981 PNF41238.1 GFWV01022530 MAA47257.1 GANP01008093 JAB76375.1 GEDV01004721 JAP83836.1 GFPF01010908 MAA22054.1 ABJB010036788 ABJB010420628 DS687722 EEC04479.1 GFWV01001475 MAA26205.1 GDIQ01095264 JAL56462.1 GDIQ01011810 JAN82927.1 GBBL01002033 JAC25287.1 AJVK01015792 GBBM01000328 JAC35090.1 GFAC01006894 JAT92294.1 GBZX01000265 JAG92475.1 GBBK01001958 JAC22524.1 GFAA01000440 JAU02995.1 GBBK01001931 JAC22551.1 GEDC01031150 JAS06148.1 LNIX01000006 OXA52993.1 GEZM01030098 JAV85809.1 UFQT01001596 SSX31102.1 UFQS01001600 UFQT01001600 SSX11553.1 SSX31120.1 GDIQ01095262 JAL56464.1 GBHO01015180 GBHO01015179 GDHC01011015 JAG28424.1 JAG28425.1 JAQ07614.1 GDIP01182255 JAJ41147.1 GEHC01001356 JAV46289.1
KPJ00554.1 NWSH01001084 PCG72709.1 KQ461108 KPJ09087.1 DS231833 EDS32248.1 GFDL01007751 JAV27294.1 CH477491 EAT40023.1 GFDF01011074 JAV03010.1 KC740768 AGM32592.1 GAPW01005057 JAC08541.1 AJWK01001776 JXUM01039167 JXUM01039168 KQ561193 KXJ79409.1 ATLV01015326 KE525006 KFB40342.1 GALA01000788 JAA94064.1 KK852598 KDR20537.1 AXCN02000544 KQ981560 KYN39850.1 AAAB01008807 EAA04310.4 GALX01001614 JAB66852.1 GL888828 EGI57730.1 APCN01000180 DS234996 EEB10164.1 KQ979878 KYN18691.1 KQ414699 KOC63429.1 KK107260 QOIP01000001 EZA54249.1 RLU26241.1 KQ976438 KYM86874.1 ADTU01012403 GGFJ01009623 MBW58764.1 GGFM01002314 MBW23065.1 GANO01004120 JAB55751.1 KQ971334 EEZ99657.2 GGFK01007392 MBW40713.1 GL438137 EFN69503.1 EU934396 ADMH02002078 ACI30174.1 ETN59499.1 GL449994 EFN81610.1 KQ435891 KOX69404.1 GBYB01000472 JAG70239.1 KQ977796 KYM99618.1 GEZM01030096 JAV85811.1 GEZM01030097 JAV85810.1 KZ288212 PBC32831.1 NNAY01001263 OXU24568.1 AXCM01004785 GDQN01009115 JAT81939.1 GDIQ01178681 JAK73044.1 GDIQ01199578 JAK52147.1 KK119091 KFM74667.1 GDIP01235055 JAI88346.1 GL732574 EFX75498.1 GDIQ01095263 JAL56463.1 APGK01026508 KB740635 KB631992 ENN79825.1 ERL87768.1 GDIP01189400 JAJ34002.1 GDIP01043165 LRGB01000872 JAM60550.1 KZS15356.1 GDIP01215855 JAJ07547.1 GDIP01121757 JAL81957.1 GDIQ01200876 JAK50849.1 GECU01028502 JAS79204.1 GECZ01029289 GECZ01027354 JAS40480.1 JAS42415.1 CVRI01000054 CRL00215.1 EU252459 ACD12008.1 GEFM01000782 JAP75014.1 GFJQ02008283 JAV98686.1 JH431809 GADI01008360 JAA65448.1 GACK01008412 JAA56622.1 NEVH01002981 PNF41238.1 GFWV01022530 MAA47257.1 GANP01008093 JAB76375.1 GEDV01004721 JAP83836.1 GFPF01010908 MAA22054.1 ABJB010036788 ABJB010420628 DS687722 EEC04479.1 GFWV01001475 MAA26205.1 GDIQ01095264 JAL56462.1 GDIQ01011810 JAN82927.1 GBBL01002033 JAC25287.1 AJVK01015792 GBBM01000328 JAC35090.1 GFAC01006894 JAT92294.1 GBZX01000265 JAG92475.1 GBBK01001958 JAC22524.1 GFAA01000440 JAU02995.1 GBBK01001931 JAC22551.1 GEDC01031150 JAS06148.1 LNIX01000006 OXA52993.1 GEZM01030098 JAV85809.1 UFQT01001596 SSX31102.1 UFQS01001600 UFQT01001600 SSX11553.1 SSX31120.1 GDIQ01095262 JAL56464.1 GBHO01015180 GBHO01015179 GDHC01011015 JAG28424.1 JAG28425.1 JAQ07614.1 GDIP01182255 JAJ41147.1 GEHC01001356 JAV46289.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000053240
UP000002320
+ More
UP000008820 UP000092461 UP000075881 UP000069940 UP000249989 UP000030765 UP000027135 UP000075886 UP000075885 UP000078541 UP000075902 UP000007062 UP000007755 UP000075840 UP000009046 UP000078492 UP000053825 UP000053097 UP000279307 UP000078540 UP000005205 UP000007266 UP000000311 UP000000673 UP000005203 UP000008237 UP000053105 UP000002358 UP000078542 UP000075901 UP000242457 UP000215335 UP000075883 UP000054359 UP000000305 UP000019118 UP000030742 UP000076858 UP000183832 UP000235965 UP000001555 UP000092462 UP000198287
UP000008820 UP000092461 UP000075881 UP000069940 UP000249989 UP000030765 UP000027135 UP000075886 UP000075885 UP000078541 UP000075902 UP000007062 UP000007755 UP000075840 UP000009046 UP000078492 UP000053825 UP000053097 UP000279307 UP000078540 UP000005205 UP000007266 UP000000311 UP000000673 UP000005203 UP000008237 UP000053105 UP000002358 UP000078542 UP000075901 UP000242457 UP000215335 UP000075883 UP000054359 UP000000305 UP000019118 UP000030742 UP000076858 UP000183832 UP000235965 UP000001555 UP000092462 UP000198287
PRIDE
Pfam
PF05434 Tmemb_9
Interpro
IPR008853
TMEM9
ProteinModelPortal
H9IXY2
A0A2W1BHP3
A0A212F9N5
A0A194Q5B8
A0A2A4JLL6
A0A194QUE4
+ More
B0W3V1 A0A1Q3FIC0 Q16ZG7 A0A1L8D9K5 R4V1N3 A0A023EI23 A0A1B0C917 A0A182JTP2 A0A182G242 A0A084VQU8 T1D4Y0 A0A067RCE1 A0A182QGI4 A0A182P232 A0A195FGP2 A0A182U862 Q7QJ43 V5GYC8 F4X7B8 A0A182I2U2 E0V9V8 A0A151J637 A0A0L7QY08 A0A026WGP3 A0A195BM69 A0A158ND58 A0A2M4C0H1 A0A2M3Z3I6 U5EEX9 D6WIJ0 A0A2M4AIW0 E2AAQ3 B6DE27 A0A088A9P6 E2BRR8 A0A0N0BCR8 A0A0C9QIM6 K7J6B4 A0A151IFD4 A0A1Y1MJD3 A0A182T1X6 A0A1Y1MM90 A0A2A3EP73 A0A232F277 A0A182M0Y9 A0A1E1W4S2 A0A0P5LDQ5 A0A0P5JRT6 A0A087UBC8 A0A0P4XYL9 E9GXY2 A0A0P5RWN6 N6UDP3 A0A0P5B5C8 A0A0P5ZIB1 A0A0P4ZAW5 A0A0P5TTI8 A0A0P5L2P6 A0A1B6HWZ5 A0A1B6ERE5 A0A1J1IIZ7 A0A0U1U058 A0A131Y6C1 A0A1Z5KUB7 T1J2U0 A0A0K8R3I8 L7LYE8 A0A2J7RK74 A0A293MNI7 V5H0A1 A0A131Z036 A0A224Z6L4 B7PD07 A0A293LHM2 A0A0P5SD31 A0A0P6HZJ0 A0A023FUE0 A0A1B0DJG7 A0A023GPA1 A0A1E1WZ69 A0A0C9R669 A0A023FMQ1 A0A1E1XUN0 A0A023FL44 A0A1B6BY24 A0A226E5T3 A0A1Y1MJ29 A0A336ML15 A0A336L3M2 A0A0P5S8I4 A0A0A9YFX8 A0A0P5BP59 A0A1W7R544
B0W3V1 A0A1Q3FIC0 Q16ZG7 A0A1L8D9K5 R4V1N3 A0A023EI23 A0A1B0C917 A0A182JTP2 A0A182G242 A0A084VQU8 T1D4Y0 A0A067RCE1 A0A182QGI4 A0A182P232 A0A195FGP2 A0A182U862 Q7QJ43 V5GYC8 F4X7B8 A0A182I2U2 E0V9V8 A0A151J637 A0A0L7QY08 A0A026WGP3 A0A195BM69 A0A158ND58 A0A2M4C0H1 A0A2M3Z3I6 U5EEX9 D6WIJ0 A0A2M4AIW0 E2AAQ3 B6DE27 A0A088A9P6 E2BRR8 A0A0N0BCR8 A0A0C9QIM6 K7J6B4 A0A151IFD4 A0A1Y1MJD3 A0A182T1X6 A0A1Y1MM90 A0A2A3EP73 A0A232F277 A0A182M0Y9 A0A1E1W4S2 A0A0P5LDQ5 A0A0P5JRT6 A0A087UBC8 A0A0P4XYL9 E9GXY2 A0A0P5RWN6 N6UDP3 A0A0P5B5C8 A0A0P5ZIB1 A0A0P4ZAW5 A0A0P5TTI8 A0A0P5L2P6 A0A1B6HWZ5 A0A1B6ERE5 A0A1J1IIZ7 A0A0U1U058 A0A131Y6C1 A0A1Z5KUB7 T1J2U0 A0A0K8R3I8 L7LYE8 A0A2J7RK74 A0A293MNI7 V5H0A1 A0A131Z036 A0A224Z6L4 B7PD07 A0A293LHM2 A0A0P5SD31 A0A0P6HZJ0 A0A023FUE0 A0A1B0DJG7 A0A023GPA1 A0A1E1WZ69 A0A0C9R669 A0A023FMQ1 A0A1E1XUN0 A0A023FL44 A0A1B6BY24 A0A226E5T3 A0A1Y1MJ29 A0A336ML15 A0A336L3M2 A0A0P5S8I4 A0A0A9YFX8 A0A0P5BP59 A0A1W7R544
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.991924
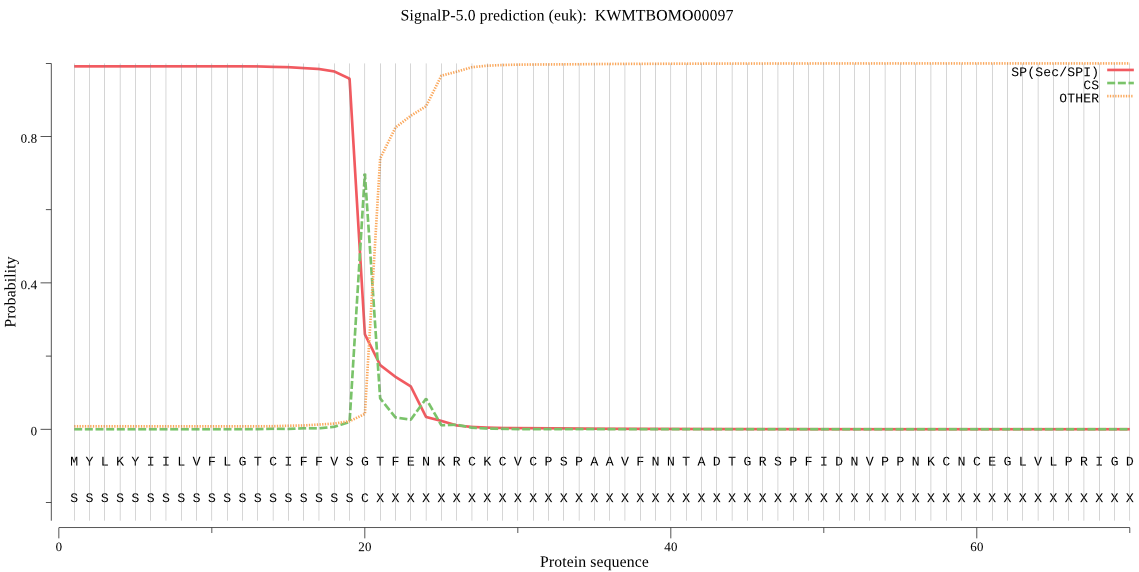
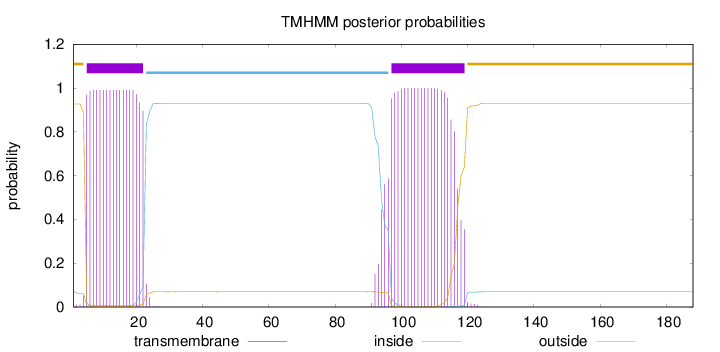
Length:
188
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
40.66984
Exp number, first 60 AAs:
17.86537
Total prob of N-in:
0.07247
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 22
inside
23 - 96
TMhelix
97 - 119
outside
120 - 188
Population Genetic Test Statistics
Pi
14.497066
Theta
16.186705
Tajima's D
-0.569998
CLR
0.09577
CSRT
0.230388480575971
Interpretation
Uncertain
