Gene
KWMTBOMO00088
Pre Gene Modal
BGIBMGA002111
Annotation
PREDICTED:_enkurin_[Papilio_xuthus]
Full name
Ribosome biogenesis protein WDR12 homolog
+ More
Enkurin
Enkurin
Location in the cell
Cytoplasmic Reliability : 1.694 Nuclear Reliability : 1.482
Sequence
CDS
ATGCTAATACTATTGCTAGTTGACACGAGAGATGGGCACATCAAGAAAATTAAGGGTTCAGGCGAAGTCCCCGAGTATTGCCTCAGACCGGATTTTGGTCGAGTTCCAGAATATTTAGTAAGGCGTAACAGAAAAATCCAGAAGGCTCTCGAAGAGATCAGGCTCGCTGATCAGAACAAGGAGAGCTTGTGCAAGCTTATCAGCGAGGAGGAACGGCAGAAGCTGTTGAAGGATTTGAAGAACAACTGGGAGCTGATGCAAAAAGCGTTTCTTCAGTTGCCGATGCTGACCGATACGATACCGAAAATTTTACGCAAAACAAAAATGGAACAAGAACTCAAGCAGCTCGAGAAAGACATCGCTTTGGTCGAGAGCAATCCTTATATCTACATTTACGAGTGA
Protein
MLILLLVDTRDGHIKKIKGSGEVPEYCLRPDFGRVPEYLVRRNRKIQKALEEIRLADQNKESLCKLISEEERQKLLKDLKNNWELMQKAFLQLPMLTDTIPKILRKTKMEQELKQLEKDIALVESNPYIYIYE
Summary
Description
Required for maturation of ribosomal RNAs and formation of the large ribosomal subunit.
Adapter that functions to localize a calcium-sensitive signal transduction machinery in sperm to a calcium-permeable ion channel.
Adapter that functions to localize a calcium-sensitive signal transduction machinery in sperm to a calcium-permeable ion channel.
Subunit
Binds calmodulin via its IQ domain. Interacts with TRPC1, TRPC2, TRPC5, but not TRPC3 (By similarity).
Similarity
Belongs to the WD repeat WDR12/YTM1 family.
Keywords
Calmodulin-binding
Cell projection
Cilium
Complete proteome
Flagellum
Reference proteome
SH3-binding
Feature
chain Enkurin
Uniprot
H9IXX8
A0A194QAU7
A0A194QUT8
A0A2H1WVK4
A0A2A4JTZ3
A0A2W1BND2
+ More
A0A212F9K2 A0A0L7LRU3 A0A182LV73 A0A182QMK2 A0A182J3C0 A0A182T795 Q17DC8 Q17DC7 A0A182G2Q3 A0A182NKR8 A0A084VQX1 A0A182JPM9 A0A1B6EWQ4 A0A1B6G4K5 A0A182YMM7 W5JUU3 A0A182WJQ3 A0A182R3H8 A0A182UYL6 A0A2C9GRR8 A0A182KQP9 Q5TWS7 A0A182U0N4 B0X8F0 A0A1Q3FSR4 A0A182XLL2 A0A1B6HX82 A0A336MZW2 A0A336LRF3 A0A1W4XIH3 A0A182FCH2 E1ZWK9 A0A1B6E8Q5 N6UEM7 U6DHF8 A0A2K5IUP3 A0A2K5W7A7 A0A2K5W7A6 A0A2K6L5S6 A0A2K6PTD6 A0A151XED2 A0A0D9RIA8 A0A195D9S7 A0A2I3RWJ5 A0A2I2Y3P2 A0A2I3HBU0 A0A087WUX1 G7N1R5 A0A2K5IUN0 A0A2K6BG27 G7PEM5 A0A2K6BG02 Q4R9A6 A0A2K6L5S9 A0A2R9BBE8 F4W4Z8 U4UIN1 A0A2K6PTE5 F6VQA4 G3ICY1 A0A091DVG4 M1EMT8 D7EI11 M3YL38 A0A286ZIE6 Q5VV23 G1S493 Q8TC29 E2RR19 A0A2J8SUS4 F7A7P7 G3QL25 A0A3Q0K142 A0A2J8MNI4 A0A2J8MNL1 A0A2K5PCN4 A0A384DQA0 A0A2K5XDA5 A0A1U7UIH1 A0A2U3W768 A0A3P4RWS3 I3MDQ7 A0A2K5MRH9 B5THN4 A0A2R9BA21 A0A2K6SED4 A0A2Y9KCT3 G5AVY5 A0A1A6H8W8 L8HZR9 E1B836 A0A2R8MNP1 A0A2Y9DSY3 A0A0N8JYV0
A0A212F9K2 A0A0L7LRU3 A0A182LV73 A0A182QMK2 A0A182J3C0 A0A182T795 Q17DC8 Q17DC7 A0A182G2Q3 A0A182NKR8 A0A084VQX1 A0A182JPM9 A0A1B6EWQ4 A0A1B6G4K5 A0A182YMM7 W5JUU3 A0A182WJQ3 A0A182R3H8 A0A182UYL6 A0A2C9GRR8 A0A182KQP9 Q5TWS7 A0A182U0N4 B0X8F0 A0A1Q3FSR4 A0A182XLL2 A0A1B6HX82 A0A336MZW2 A0A336LRF3 A0A1W4XIH3 A0A182FCH2 E1ZWK9 A0A1B6E8Q5 N6UEM7 U6DHF8 A0A2K5IUP3 A0A2K5W7A7 A0A2K5W7A6 A0A2K6L5S6 A0A2K6PTD6 A0A151XED2 A0A0D9RIA8 A0A195D9S7 A0A2I3RWJ5 A0A2I2Y3P2 A0A2I3HBU0 A0A087WUX1 G7N1R5 A0A2K5IUN0 A0A2K6BG27 G7PEM5 A0A2K6BG02 Q4R9A6 A0A2K6L5S9 A0A2R9BBE8 F4W4Z8 U4UIN1 A0A2K6PTE5 F6VQA4 G3ICY1 A0A091DVG4 M1EMT8 D7EI11 M3YL38 A0A286ZIE6 Q5VV23 G1S493 Q8TC29 E2RR19 A0A2J8SUS4 F7A7P7 G3QL25 A0A3Q0K142 A0A2J8MNI4 A0A2J8MNL1 A0A2K5PCN4 A0A384DQA0 A0A2K5XDA5 A0A1U7UIH1 A0A2U3W768 A0A3P4RWS3 I3MDQ7 A0A2K5MRH9 B5THN4 A0A2R9BA21 A0A2K6SED4 A0A2Y9KCT3 G5AVY5 A0A1A6H8W8 L8HZR9 E1B836 A0A2R8MNP1 A0A2Y9DSY3 A0A0N8JYV0
Pubmed
19121390
26354079
28756777
22118469
26227816
17510324
+ More
26483478 24438588 25244985 20920257 23761445 20966253 12364791 14747013 17210077 20798317 23537049 25362486 16136131 15164054 22002653 15944441 22722832 21719571 19892987 21804562 23236062 18362917 19820115 11181995 15385169 14702039 15489334 16341006 12481130 15114417 24813606 21993625 22751099 19393038
26483478 24438588 25244985 20920257 23761445 20966253 12364791 14747013 17210077 20798317 23537049 25362486 16136131 15164054 22002653 15944441 22722832 21719571 19892987 21804562 23236062 18362917 19820115 11181995 15385169 14702039 15489334 16341006 12481130 15114417 24813606 21993625 22751099 19393038
EMBL
BABH01015783
BABH01015784
KQ459463
KPJ00546.1
KQ461108
KPJ09079.1
+ More
ODYU01011323 SOQ56996.1 NWSH01000586 PCG75497.1 KZ149952 PZC76572.1 AGBW02009564 OWR50426.1 JTDY01000228 KOB78188.1 AXCM01002371 AXCN02000542 CH477297 EAT44362.1 EAT44363.1 JXUM01039933 JXUM01039934 KQ561224 KXJ79302.1 ATLV01015338 KE525006 KFB40365.1 GECZ01027323 JAS42446.1 GECZ01012394 JAS57375.1 ADMH02000008 ETN68122.1 APCN01000179 AAAB01008807 EAL41843.3 DS232486 EDS42486.1 GFDL01004394 JAV30651.1 GECU01028461 JAS79245.1 UFQT01004512 SSX35710.1 UFQS01000091 UFQT01000091 SSW99149.1 SSX19531.1 GL434853 EFN74446.1 GEDC01003002 JAS34296.1 APGK01038383 KB740954 ENN77092.1 HAAF01009743 CCP81567.1 AQIA01070383 AQIA01070384 AQIA01070385 AQIA01070386 AQIA01070387 AQIA01070388 AQIA01070389 KQ982254 KYQ58707.1 AQIB01160473 KQ981082 KYN09633.1 AACZ04070749 AACZ04070750 AACZ04070751 NBAG03000250 PNI61085.1 CABD030069631 CABD030069632 CABD030069633 CABD030069634 CABD030069635 CABD030069636 CABD030069637 CABD030069638 CABD030069639 CABD030069640 ADFV01049909 ADFV01049910 ADFV01049911 ADFV01049912 AL512598 CM001261 EHH18942.1 CM001284 EHH64619.1 AB168190 BAE00315.1 AJFE02073497 AJFE02073498 AJFE02073499 GL887596 EGI70694.1 KB632338 ERL92897.1 JH001974 EGW14195.1 KN121889 KFO35087.1 JP009327 AER97924.1 DS497669 EFA12927.1 AEYP01043676 AEYP01043677 AEMK02000074 CH471072 EAW86117.1 AY454125 AK095021 AK292495 BC026165 AAEX03001175 NDHI03003541 PNJ24515.1 EAAA01000006 PNI61086.1 PNI61087.1 PNI61088.1 CYRY02043751 VCX38371.1 AGTP01016586 EU939760 ACH73242.1 JH167180 EHB01196.1 LZPO01044445 OBS74305.1 JH882436 ELR49418.1 JARO02004982 KPP67553.1
ODYU01011323 SOQ56996.1 NWSH01000586 PCG75497.1 KZ149952 PZC76572.1 AGBW02009564 OWR50426.1 JTDY01000228 KOB78188.1 AXCM01002371 AXCN02000542 CH477297 EAT44362.1 EAT44363.1 JXUM01039933 JXUM01039934 KQ561224 KXJ79302.1 ATLV01015338 KE525006 KFB40365.1 GECZ01027323 JAS42446.1 GECZ01012394 JAS57375.1 ADMH02000008 ETN68122.1 APCN01000179 AAAB01008807 EAL41843.3 DS232486 EDS42486.1 GFDL01004394 JAV30651.1 GECU01028461 JAS79245.1 UFQT01004512 SSX35710.1 UFQS01000091 UFQT01000091 SSW99149.1 SSX19531.1 GL434853 EFN74446.1 GEDC01003002 JAS34296.1 APGK01038383 KB740954 ENN77092.1 HAAF01009743 CCP81567.1 AQIA01070383 AQIA01070384 AQIA01070385 AQIA01070386 AQIA01070387 AQIA01070388 AQIA01070389 KQ982254 KYQ58707.1 AQIB01160473 KQ981082 KYN09633.1 AACZ04070749 AACZ04070750 AACZ04070751 NBAG03000250 PNI61085.1 CABD030069631 CABD030069632 CABD030069633 CABD030069634 CABD030069635 CABD030069636 CABD030069637 CABD030069638 CABD030069639 CABD030069640 ADFV01049909 ADFV01049910 ADFV01049911 ADFV01049912 AL512598 CM001261 EHH18942.1 CM001284 EHH64619.1 AB168190 BAE00315.1 AJFE02073497 AJFE02073498 AJFE02073499 GL887596 EGI70694.1 KB632338 ERL92897.1 JH001974 EGW14195.1 KN121889 KFO35087.1 JP009327 AER97924.1 DS497669 EFA12927.1 AEYP01043676 AEYP01043677 AEMK02000074 CH471072 EAW86117.1 AY454125 AK095021 AK292495 BC026165 AAEX03001175 NDHI03003541 PNJ24515.1 EAAA01000006 PNI61086.1 PNI61087.1 PNI61088.1 CYRY02043751 VCX38371.1 AGTP01016586 EU939760 ACH73242.1 JH167180 EHB01196.1 LZPO01044445 OBS74305.1 JH882436 ELR49418.1 JARO02004982 KPP67553.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000037510
+ More
UP000075883 UP000075886 UP000075880 UP000075901 UP000008820 UP000069940 UP000249989 UP000075884 UP000030765 UP000075881 UP000076408 UP000000673 UP000075920 UP000075900 UP000075903 UP000075840 UP000075882 UP000007062 UP000075902 UP000002320 UP000076407 UP000192223 UP000069272 UP000000311 UP000019118 UP000233080 UP000233180 UP000233200 UP000075809 UP000029965 UP000078492 UP000002277 UP000001519 UP000001073 UP000005640 UP000233120 UP000009130 UP000240080 UP000007755 UP000030742 UP000002281 UP000001075 UP000028990 UP000007266 UP000000715 UP000008227 UP000002254 UP000008144 UP000233040 UP000261680 UP000291021 UP000233140 UP000189704 UP000245340 UP000005215 UP000233060 UP000233220 UP000248482 UP000006813 UP000092124 UP000009136 UP000008225 UP000248480 UP000034805 UP000192224
UP000075883 UP000075886 UP000075880 UP000075901 UP000008820 UP000069940 UP000249989 UP000075884 UP000030765 UP000075881 UP000076408 UP000000673 UP000075920 UP000075900 UP000075903 UP000075840 UP000075882 UP000007062 UP000075902 UP000002320 UP000076407 UP000192223 UP000069272 UP000000311 UP000019118 UP000233080 UP000233180 UP000233200 UP000075809 UP000029965 UP000078492 UP000002277 UP000001519 UP000001073 UP000005640 UP000233120 UP000009130 UP000240080 UP000007755 UP000030742 UP000002281 UP000001075 UP000028990 UP000007266 UP000000715 UP000008227 UP000002254 UP000008144 UP000233040 UP000261680 UP000291021 UP000233140 UP000189704 UP000245340 UP000005215 UP000233060 UP000233220 UP000248482 UP000006813 UP000092124 UP000009136 UP000008225 UP000248480 UP000034805 UP000192224
Pfam
Interpro
IPR026150
Enkur
+ More
IPR027012 Enkurin_dom
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR000086 NUDIX_hydrolase_dom
IPR016706 Cleav_polyA_spec_factor_su5
IPR028599 WDR12/Ytm1
IPR003959 ATPase_AAA_core
IPR012972 NLE
IPR020472 G-protein_beta_WD-40_rep
IPR001163 LSM_dom_euk/arc
IPR036322 WD40_repeat_dom_sf
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR019775 WD40_repeat_CS
IPR032501 Prot_ATP_ID_OB
IPR010920 LSM_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR034102 Sm_D1
IPR003593 AAA+_ATPase
IPR041569 AAA_lid_3
IPR001680 WD40_repeat
IPR005937 26S_Psome_P45-like
IPR017986 WD40_repeat_dom
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR027012 Enkurin_dom
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR000086 NUDIX_hydrolase_dom
IPR016706 Cleav_polyA_spec_factor_su5
IPR028599 WDR12/Ytm1
IPR003959 ATPase_AAA_core
IPR012972 NLE
IPR020472 G-protein_beta_WD-40_rep
IPR001163 LSM_dom_euk/arc
IPR036322 WD40_repeat_dom_sf
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR019775 WD40_repeat_CS
IPR032501 Prot_ATP_ID_OB
IPR010920 LSM_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR034102 Sm_D1
IPR003593 AAA+_ATPase
IPR041569 AAA_lid_3
IPR001680 WD40_repeat
IPR005937 26S_Psome_P45-like
IPR017986 WD40_repeat_dom
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9IXX8
A0A194QAU7
A0A194QUT8
A0A2H1WVK4
A0A2A4JTZ3
A0A2W1BND2
+ More
A0A212F9K2 A0A0L7LRU3 A0A182LV73 A0A182QMK2 A0A182J3C0 A0A182T795 Q17DC8 Q17DC7 A0A182G2Q3 A0A182NKR8 A0A084VQX1 A0A182JPM9 A0A1B6EWQ4 A0A1B6G4K5 A0A182YMM7 W5JUU3 A0A182WJQ3 A0A182R3H8 A0A182UYL6 A0A2C9GRR8 A0A182KQP9 Q5TWS7 A0A182U0N4 B0X8F0 A0A1Q3FSR4 A0A182XLL2 A0A1B6HX82 A0A336MZW2 A0A336LRF3 A0A1W4XIH3 A0A182FCH2 E1ZWK9 A0A1B6E8Q5 N6UEM7 U6DHF8 A0A2K5IUP3 A0A2K5W7A7 A0A2K5W7A6 A0A2K6L5S6 A0A2K6PTD6 A0A151XED2 A0A0D9RIA8 A0A195D9S7 A0A2I3RWJ5 A0A2I2Y3P2 A0A2I3HBU0 A0A087WUX1 G7N1R5 A0A2K5IUN0 A0A2K6BG27 G7PEM5 A0A2K6BG02 Q4R9A6 A0A2K6L5S9 A0A2R9BBE8 F4W4Z8 U4UIN1 A0A2K6PTE5 F6VQA4 G3ICY1 A0A091DVG4 M1EMT8 D7EI11 M3YL38 A0A286ZIE6 Q5VV23 G1S493 Q8TC29 E2RR19 A0A2J8SUS4 F7A7P7 G3QL25 A0A3Q0K142 A0A2J8MNI4 A0A2J8MNL1 A0A2K5PCN4 A0A384DQA0 A0A2K5XDA5 A0A1U7UIH1 A0A2U3W768 A0A3P4RWS3 I3MDQ7 A0A2K5MRH9 B5THN4 A0A2R9BA21 A0A2K6SED4 A0A2Y9KCT3 G5AVY5 A0A1A6H8W8 L8HZR9 E1B836 A0A2R8MNP1 A0A2Y9DSY3 A0A0N8JYV0
A0A212F9K2 A0A0L7LRU3 A0A182LV73 A0A182QMK2 A0A182J3C0 A0A182T795 Q17DC8 Q17DC7 A0A182G2Q3 A0A182NKR8 A0A084VQX1 A0A182JPM9 A0A1B6EWQ4 A0A1B6G4K5 A0A182YMM7 W5JUU3 A0A182WJQ3 A0A182R3H8 A0A182UYL6 A0A2C9GRR8 A0A182KQP9 Q5TWS7 A0A182U0N4 B0X8F0 A0A1Q3FSR4 A0A182XLL2 A0A1B6HX82 A0A336MZW2 A0A336LRF3 A0A1W4XIH3 A0A182FCH2 E1ZWK9 A0A1B6E8Q5 N6UEM7 U6DHF8 A0A2K5IUP3 A0A2K5W7A7 A0A2K5W7A6 A0A2K6L5S6 A0A2K6PTD6 A0A151XED2 A0A0D9RIA8 A0A195D9S7 A0A2I3RWJ5 A0A2I2Y3P2 A0A2I3HBU0 A0A087WUX1 G7N1R5 A0A2K5IUN0 A0A2K6BG27 G7PEM5 A0A2K6BG02 Q4R9A6 A0A2K6L5S9 A0A2R9BBE8 F4W4Z8 U4UIN1 A0A2K6PTE5 F6VQA4 G3ICY1 A0A091DVG4 M1EMT8 D7EI11 M3YL38 A0A286ZIE6 Q5VV23 G1S493 Q8TC29 E2RR19 A0A2J8SUS4 F7A7P7 G3QL25 A0A3Q0K142 A0A2J8MNI4 A0A2J8MNL1 A0A2K5PCN4 A0A384DQA0 A0A2K5XDA5 A0A1U7UIH1 A0A2U3W768 A0A3P4RWS3 I3MDQ7 A0A2K5MRH9 B5THN4 A0A2R9BA21 A0A2K6SED4 A0A2Y9KCT3 G5AVY5 A0A1A6H8W8 L8HZR9 E1B836 A0A2R8MNP1 A0A2Y9DSY3 A0A0N8JYV0
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Nucleolus
Nucleoplasm
Nucleolus
Nucleoplasm
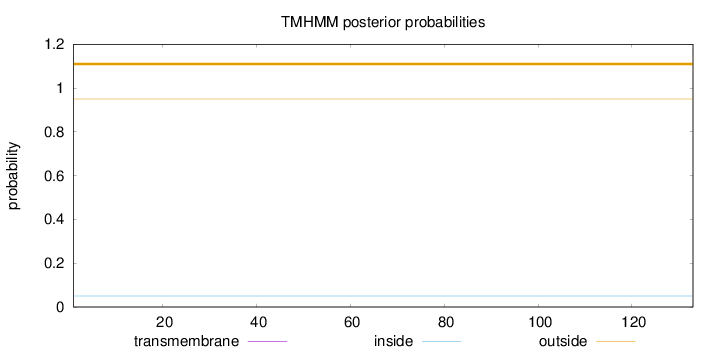
Length:
133
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00039
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04977
outside
1 - 133
Population Genetic Test Statistics
Pi
5.202077
Theta
26.316563
Tajima's D
-2.026876
CLR
0.150524
CSRT
0.0130993450327484
Interpretation
Uncertain
