Gene
KWMTBOMO00086
Pre Gene Modal
BGIBMGA002020
Annotation
PREDICTED:_pyruvate_dehydrogenase_phosphatase_regulatory_subunit?_mitochondrial_isoform_X1_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.912 Mitochondrial Reliability : 1.312
Sequence
CDS
ATGTTGGGAGGCTGTCTGAAAGGTGGCATTGACAAGTGTAATTTGTATAGAGTTGCCGGTGTCCTCAATAGGCGGTTTTCGAGCCGCTTAGATGCATTGGACTACGAAGAAAAATTAGAAGATTGCTTGTCAGTACTACCATCCAAAGCAAAGGTGGTAATATGCGGCGGGGGTGTAATGGGGGCGGCTGTCGCGTACCACCTGGCCAACCGGGGGTGGGGTGACAGAACCGTAGTCGTCGAGAAAGAAAAAGTGGGTGCAGGGAGTCGCTGGCATTCATCAGGCCTTGTTGGAGCATTCAAGCCAACACTTGCTCAGGTGAGGCTTGCCCAGTCCTCAATTAGGCTTCTTAAGGAGCTTGAGGCTCGTGGTAGGCCCACAGGGTGGAAACAATGTGGTTCACTATTATTGGCAAGAACAAGAGACCGCATGACTGTTTACAGAAGAATGAAAAGTCAGTCTGTATCGTGGAGCATAGATTGTGACTTAGTTACACCAAAGAAATGTCATGAATTATTTCCAATGCTGAATGTTGAAGATGTGCTTGGAGGCCTGTGGATTCCAGGTGATGGTGTAGGAGACCCGCATTTGCTCTGTATGTCTCTTATGAGAGAAGCAACTGATAAAGGTGTGGGTGTGATGGAAGATTGTTCTGTGACAGCTGTGCTATCGAAAGACGACAAAGTGTCTGGAGTGGAAACTACAAATGGTGCAATCGAGTGTGATTACTTTATCAACTGTGCGGGCTTTTGGGCTAGACAGGTCGGACAGTTGGCACGACCGCAGGTGAAAGTACCCTTACTACCATGTGAACATTACTATCTGCATACCAAGCCCATTGATAATCTTGATCCAATGACTCCAGTCATCCGAGACCCAGATGGTTATATATATCTTCGAGAAAGAGATGGATGCATTTTAGCAGGTGGATTCGAACCAATTGCGAAACCTGTTTATGAGGAAGAAATCGAGAACGCGTCACAACGTTGTTTGCCCGAAGACTGGGATCACTTCCACGTGTTGCTGCAGGAGTTGCTGCAACGGGTGCCCGGTCTGAACCAGGCCGTGCTCCACAAGCTGTGCAATGGACTAGAGGCATTCTCTCCTGACTGCAAGTGGATCGTTGGAGAAGCACCAGAGATGTTGAACTACCACGTGGCGGCCGGCATGAAGACTGTGGGCATATCCGCTGCCGGTGGCGTCGCTGAGGCGACGGTTGACGAGATCATCGACGGCTACAGCAAGTACGATATGCATGAGTTGGGCGTCAATCGTTTCCTTGGACTGCACAACAACAAGAGATTCCTTAGAGACCGCGTTAAGGAGGTTCCAGGCGTTCATTACGGCCTTCCGTATCCGTTCTACGAATTCGAGACTGGTCGTAACTTGCGCTTGTCTCCCATCTACCCGACACTGAGAGACAACGGAGCAGTCTTCGGTCAGGTTATGGGCTACGAGAGACCAACGTGGTTCGAGACCGTAGAGAATGAGAGCGAGAAACCGCGTCCGTTCAAGATAGCGCACACGAGAACTTTCGGGAAGCCGCCCTGGTTCGACGCGGTGCAGCGTGAGTACTGGGCGTGCCGGGAGCGGGTCGGCCTCTCCGACTACTCCTCCTTCACCAAGATAGACATACAGTCTCAAGGTCGCGAGGTGGTGGAACTCCTGCAGTATCTCTGTTCCAACGACGTGGACGTTCCCGTCGGGAGTATCATTCACACTGGAATGCAGAACGAGCGAGGGGGTTACGAGAATGACTGCAGCCTGGCTAGGATATCGGAGAACCATTACATGATGATAGCTCCGACGATCCAGCAGACGCGCTGCAAGGTGTGGCTGAAGCGACACCTGCCCAGCAATGGTTCGGTGACCCTCTCCGACGTGACGTCGATGTACACCGCCATCTGCGTCATGGGTCCGTTCACGAGGAGTTTGCTCTCCGAACTGACCGACACTGACCTGTCGCCTTCGAACTTTCCGTTCTTCACGTTCAAGGAGATCGACGTCGGTCTGGCCAATGGGATTCGGGCGATGAATCTGACGCACACCGGAGAGCTGGGATATGTGCTCTATATACCTAACGAGTTTGCTCTCCACGTGTACAATCGCCTGATGACTGTGGGTGAGAAGTACGGGATAAGCCACGTCGGTTATTATGCGTCGCGTGCGCTACGCGTCGAAAAGTTCTTCGCTTTTTGGGGCCAGGATCTCGATACGATGACGACTCCTCTAGAATGTGGACGCACGTGGCGTGTTAAATTCGACAAAGACATAAAGTTCATCGGTCGGGATGCCCTCCTGAAGCAGCGGGAGGACGGTATCCGACGTCAATATGTCCAATTGTTGTTGACCGACCACGACCACGAGCTGGACCTTTGGTCGTGGGGCGGGGAGCCGATATATAGAGACGGGAACTATTGTGGTCAGACAACCACCACCAGTTACGGATTCACATTCAAGAAACAGGTATGCCTTGGTTTCGTCGAGAAACGCGATAAGGACGGTGTTACGCAGAAAGTTGACAACGATTACGTTCTGAGCGGCCATTATGAGATCGACATAGCCGGGATAAGGTACGCGGCAAAAGTAAATCTGCACTCCCCGAACTTACCAACGAAGTATCCCGACAAGGAACGCGACGTCTACCAGGCAACGAGGAAACTGCACGAGCCCCAACAGTTCCTCGGCCGGCATTACCAGCCGTAA
Protein
MLGGCLKGGIDKCNLYRVAGVLNRRFSSRLDALDYEEKLEDCLSVLPSKAKVVICGGGVMGAAVAYHLANRGWGDRTVVVEKEKVGAGSRWHSSGLVGAFKPTLAQVRLAQSSIRLLKELEARGRPTGWKQCGSLLLARTRDRMTVYRRMKSQSVSWSIDCDLVTPKKCHELFPMLNVEDVLGGLWIPGDGVGDPHLLCMSLMREATDKGVGVMEDCSVTAVLSKDDKVSGVETTNGAIECDYFINCAGFWARQVGQLARPQVKVPLLPCEHYYLHTKPIDNLDPMTPVIRDPDGYIYLRERDGCILAGGFEPIAKPVYEEEIENASQRCLPEDWDHFHVLLQELLQRVPGLNQAVLHKLCNGLEAFSPDCKWIVGEAPEMLNYHVAAGMKTVGISAAGGVAEATVDEIIDGYSKYDMHELGVNRFLGLHNNKRFLRDRVKEVPGVHYGLPYPFYEFETGRNLRLSPIYPTLRDNGAVFGQVMGYERPTWFETVENESEKPRPFKIAHTRTFGKPPWFDAVQREYWACRERVGLSDYSSFTKIDIQSQGREVVELLQYLCSNDVDVPVGSIIHTGMQNERGGYENDCSLARISENHYMMIAPTIQQTRCKVWLKRHLPSNGSVTLSDVTSMYTAICVMGPFTRSLLSELTDTDLSPSNFPFFTFKEIDVGLANGIRAMNLTHTGELGYVLYIPNEFALHVYNRLMTVGEKYGISHVGYYASRALRVEKFFAFWGQDLDTMTTPLECGRTWRVKFDKDIKFIGRDALLKQREDGIRRQYVQLLLTDHDHELDLWSWGGEPIYRDGNYCGQTTTTSYGFTFKKQVCLGFVEKRDKDGVTQKVDNDYVLSGHYEIDIAGIRYAAKVNLHSPNLPTKYPDKERDVYQATRKLHEPQQFLGRHYQP
Summary
Uniprot
A0A2W1BXT2
A0A194QU73
A0A212F9L9
A0A2J7QWV3
A0A2J7QWU2
D6WGA9
+ More
N6T053 E2C0J5 E2AFR4 A0A0J7L4H8 A0A1Q3FF35 A0A1Q3FFS8 A0A023F461 A0A182R2U8 A0A1Q3FFL9 A0A158NAM6 A0A1S4FMS9 A0A182WAU6 Q16VJ0 A0A182Q1A7 A0A023EWM8 A0A026WLF6 A0A224X726 A0A195DQN8 A0A195CLD2 A0A182NN92 A0A182IKB4 A0A151XIH2 Q7Q2M2 F4W6V4 A0A182LPU8 A0A182G1Y4 A0A067R5Z2 A0A182K281 A0A151I694 A0A2A3EJU0 A0A182HGJ4 A0A3L8DCT8 A0A182YI71 J9K0C5 B0XBZ6 A0A232EST7 A0A182PNG5 A0A084W1B7 A0A2M4CVA4 A0A2M4AI98 A0A2J7QWU4 A0A2S2QJN3 A0A0L7QZ50 A0A2H8TN07 A0A1B6D3W9 E9J3K1 U5EU87 A0A1B6FLK8 A0A154PB33 A0A2H1V324 A0A1B6KYV5 A0A1B6L0J7 E0VHC3 A0A1Y1K630 A0A0A9WV29 A0A146L4D4 T1PCJ5 A0A1I8M5W6 A0A182FLS3 A0A195EVI8 A0A1B0C9I4 W8B260 A0A034VKT0 A0A0K8U533 A0A1L8E2H1 A0A1I8PX73 B4M7A7 B4L5R6 A0A0L0BTF1 B3MXF7 A0A3B0KRM2 Q29J50 B4NP81 A0A0M4ETX9 A0A1A9VCK3 B4R4H6 A0A1B0GA17 Q9W4K8 A0A1W4V232 A0A1A9ZEI2 A0A1A9XBV0 A0A1B0BE01 B4Q063 B3NU25 A0A1A9WT20 Q8MQN6 B4JKC8 T1IHI3 B4I0Z6 A0A0P5RUW1 A0A0P6HC22 A0A0P6FBP3 A0A0P5S5F8
N6T053 E2C0J5 E2AFR4 A0A0J7L4H8 A0A1Q3FF35 A0A1Q3FFS8 A0A023F461 A0A182R2U8 A0A1Q3FFL9 A0A158NAM6 A0A1S4FMS9 A0A182WAU6 Q16VJ0 A0A182Q1A7 A0A023EWM8 A0A026WLF6 A0A224X726 A0A195DQN8 A0A195CLD2 A0A182NN92 A0A182IKB4 A0A151XIH2 Q7Q2M2 F4W6V4 A0A182LPU8 A0A182G1Y4 A0A067R5Z2 A0A182K281 A0A151I694 A0A2A3EJU0 A0A182HGJ4 A0A3L8DCT8 A0A182YI71 J9K0C5 B0XBZ6 A0A232EST7 A0A182PNG5 A0A084W1B7 A0A2M4CVA4 A0A2M4AI98 A0A2J7QWU4 A0A2S2QJN3 A0A0L7QZ50 A0A2H8TN07 A0A1B6D3W9 E9J3K1 U5EU87 A0A1B6FLK8 A0A154PB33 A0A2H1V324 A0A1B6KYV5 A0A1B6L0J7 E0VHC3 A0A1Y1K630 A0A0A9WV29 A0A146L4D4 T1PCJ5 A0A1I8M5W6 A0A182FLS3 A0A195EVI8 A0A1B0C9I4 W8B260 A0A034VKT0 A0A0K8U533 A0A1L8E2H1 A0A1I8PX73 B4M7A7 B4L5R6 A0A0L0BTF1 B3MXF7 A0A3B0KRM2 Q29J50 B4NP81 A0A0M4ETX9 A0A1A9VCK3 B4R4H6 A0A1B0GA17 Q9W4K8 A0A1W4V232 A0A1A9ZEI2 A0A1A9XBV0 A0A1B0BE01 B4Q063 B3NU25 A0A1A9WT20 Q8MQN6 B4JKC8 T1IHI3 B4I0Z6 A0A0P5RUW1 A0A0P6HC22 A0A0P6FBP3 A0A0P5S5F8
Pubmed
28756777
26354079
22118469
18362917
19820115
23537049
+ More
20798317 25474469 21347285 17510324 24945155 24508170 12364791 14747013 17210077 21719571 20966253 26483478 24845553 30249741 25244985 28648823 24438588 21282665 20566863 28004739 25401762 26823975 25315136 24495485 25348373 17994087 26108605 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
20798317 25474469 21347285 17510324 24945155 24508170 12364791 14747013 17210077 21719571 20966253 26483478 24845553 30249741 25244985 28648823 24438588 21282665 20566863 28004739 25401762 26823975 25315136 24495485 25348373 17994087 26108605 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
EMBL
KZ149952
PZC76573.1
KQ461108
KPJ09078.1
AGBW02009564
OWR50427.1
+ More
NEVH01009418 PNF33055.1 PNF33057.1 KQ971329 EFA01139.2 APGK01057365 APGK01057366 KB741280 KB631604 ENN70883.1 ERL84603.1 GL451800 EFN78512.1 GL439124 EFN67725.1 LBMM01000672 KMQ97817.1 GFDL01008881 JAV26164.1 GFDL01008636 JAV26409.1 GBBI01002441 JAC16271.1 GFDL01008705 JAV26340.1 ADTU01010381 ADTU01010382 CH477590 EAT38581.1 AXCN02001901 GAPW01000141 JAC13457.1 KK107167 EZA56486.1 GFTR01008171 JAW08255.1 KQ980612 KYN15161.1 KQ977642 KYN00894.1 KQ982108 KYQ60010.1 AAAB01008968 EAA13207.3 GL887762 EGI70067.1 JXUM01039375 JXUM01039376 KQ561202 KXJ79372.1 KK852680 KDR18611.1 KQ976413 KYM90356.1 KZ288223 PBC31960.1 APCN01006423 APCN01006424 APCN01006425 QOIP01000010 RLU18141.1 ABLF02040040 DS232664 EDS44559.1 NNAY01002374 OXU21423.1 ATLV01019258 KE525266 KFB44011.1 GGFL01005099 MBW69277.1 GGFK01007146 MBW40467.1 PNF33058.1 GGMS01008527 MBY77730.1 KQ414683 KOC63890.1 GFXV01003738 MBW15543.1 GEDC01016914 JAS20384.1 GL768066 EFZ12581.1 GANO01002432 JAB57439.1 GECZ01018705 JAS51064.1 KQ434846 KZC08438.1 ODYU01000239 SOQ34684.1 GEBQ01023350 GEBQ01016099 JAT16627.1 JAT23878.1 GEBQ01029402 GEBQ01022802 JAT10575.1 JAT17175.1 DS235165 EEB12779.1 GEZM01091335 JAV56902.1 GBHO01033236 GDHC01014404 JAG10368.1 JAQ04225.1 GDHC01016513 JAQ02116.1 KA645865 AFP60494.1 KQ981954 KYN32275.1 AJWK01002378 GAMC01013968 JAB92587.1 GAKP01016522 JAC42430.1 GDHF01031965 GDHF01030562 GDHF01023846 GDHF01022617 GDHF01011659 GDHF01009361 JAI20349.1 JAI21752.1 JAI28468.1 JAI29697.1 JAI40655.1 JAI42953.1 GFDF01001173 JAV12911.1 CH940653 EDW62674.1 CH933811 EDW06525.1 JRES01001379 KNC23268.1 CH902630 EDV38422.1 OUUW01000019 SPP89299.1 CH379063 EAL32452.1 KRT06028.1 CH964291 EDW86321.2 CP012528 ALC48435.1 CM000366 EDX17066.1 CCAG010018798 AE014298 BT150297 AAF45943.3 AGW24079.1 JXJN01012736 CM000162 EDX01215.1 CH954180 EDV45801.1 AY128477 AAM75070.1 CH916370 EDW00031.1 JH429910 CH480819 EDW53177.1 GDIQ01096329 JAL55397.1 GDIQ01022313 JAN72424.1 GDIQ01062449 JAN32288.1 GDIQ01242279 GDIQ01188644 GDIQ01132234 GDIQ01092191 GDIQ01063992 GDIQ01060501 JAL59535.1
NEVH01009418 PNF33055.1 PNF33057.1 KQ971329 EFA01139.2 APGK01057365 APGK01057366 KB741280 KB631604 ENN70883.1 ERL84603.1 GL451800 EFN78512.1 GL439124 EFN67725.1 LBMM01000672 KMQ97817.1 GFDL01008881 JAV26164.1 GFDL01008636 JAV26409.1 GBBI01002441 JAC16271.1 GFDL01008705 JAV26340.1 ADTU01010381 ADTU01010382 CH477590 EAT38581.1 AXCN02001901 GAPW01000141 JAC13457.1 KK107167 EZA56486.1 GFTR01008171 JAW08255.1 KQ980612 KYN15161.1 KQ977642 KYN00894.1 KQ982108 KYQ60010.1 AAAB01008968 EAA13207.3 GL887762 EGI70067.1 JXUM01039375 JXUM01039376 KQ561202 KXJ79372.1 KK852680 KDR18611.1 KQ976413 KYM90356.1 KZ288223 PBC31960.1 APCN01006423 APCN01006424 APCN01006425 QOIP01000010 RLU18141.1 ABLF02040040 DS232664 EDS44559.1 NNAY01002374 OXU21423.1 ATLV01019258 KE525266 KFB44011.1 GGFL01005099 MBW69277.1 GGFK01007146 MBW40467.1 PNF33058.1 GGMS01008527 MBY77730.1 KQ414683 KOC63890.1 GFXV01003738 MBW15543.1 GEDC01016914 JAS20384.1 GL768066 EFZ12581.1 GANO01002432 JAB57439.1 GECZ01018705 JAS51064.1 KQ434846 KZC08438.1 ODYU01000239 SOQ34684.1 GEBQ01023350 GEBQ01016099 JAT16627.1 JAT23878.1 GEBQ01029402 GEBQ01022802 JAT10575.1 JAT17175.1 DS235165 EEB12779.1 GEZM01091335 JAV56902.1 GBHO01033236 GDHC01014404 JAG10368.1 JAQ04225.1 GDHC01016513 JAQ02116.1 KA645865 AFP60494.1 KQ981954 KYN32275.1 AJWK01002378 GAMC01013968 JAB92587.1 GAKP01016522 JAC42430.1 GDHF01031965 GDHF01030562 GDHF01023846 GDHF01022617 GDHF01011659 GDHF01009361 JAI20349.1 JAI21752.1 JAI28468.1 JAI29697.1 JAI40655.1 JAI42953.1 GFDF01001173 JAV12911.1 CH940653 EDW62674.1 CH933811 EDW06525.1 JRES01001379 KNC23268.1 CH902630 EDV38422.1 OUUW01000019 SPP89299.1 CH379063 EAL32452.1 KRT06028.1 CH964291 EDW86321.2 CP012528 ALC48435.1 CM000366 EDX17066.1 CCAG010018798 AE014298 BT150297 AAF45943.3 AGW24079.1 JXJN01012736 CM000162 EDX01215.1 CH954180 EDV45801.1 AY128477 AAM75070.1 CH916370 EDW00031.1 JH429910 CH480819 EDW53177.1 GDIQ01096329 JAL55397.1 GDIQ01022313 JAN72424.1 GDIQ01062449 JAN32288.1 GDIQ01242279 GDIQ01188644 GDIQ01132234 GDIQ01092191 GDIQ01063992 GDIQ01060501 JAL59535.1
Proteomes
UP000053240
UP000007151
UP000235965
UP000007266
UP000019118
UP000030742
+ More
UP000008237 UP000000311 UP000036403 UP000075900 UP000005205 UP000075920 UP000008820 UP000075886 UP000053097 UP000078492 UP000078542 UP000075884 UP000075880 UP000075809 UP000007062 UP000007755 UP000075882 UP000069940 UP000249989 UP000027135 UP000075881 UP000078540 UP000242457 UP000075840 UP000279307 UP000076408 UP000007819 UP000002320 UP000215335 UP000075885 UP000030765 UP000053825 UP000076502 UP000009046 UP000095301 UP000069272 UP000078541 UP000092461 UP000095300 UP000008792 UP000009192 UP000037069 UP000007801 UP000268350 UP000001819 UP000007798 UP000092553 UP000078200 UP000000304 UP000092444 UP000000803 UP000192221 UP000092445 UP000092443 UP000092460 UP000002282 UP000008711 UP000091820 UP000001070 UP000001292
UP000008237 UP000000311 UP000036403 UP000075900 UP000005205 UP000075920 UP000008820 UP000075886 UP000053097 UP000078492 UP000078542 UP000075884 UP000075880 UP000075809 UP000007062 UP000007755 UP000075882 UP000069940 UP000249989 UP000027135 UP000075881 UP000078540 UP000242457 UP000075840 UP000279307 UP000076408 UP000007819 UP000002320 UP000215335 UP000075885 UP000030765 UP000053825 UP000076502 UP000009046 UP000095301 UP000069272 UP000078541 UP000092461 UP000095300 UP000008792 UP000009192 UP000037069 UP000007801 UP000268350 UP000001819 UP000007798 UP000092553 UP000078200 UP000000304 UP000092444 UP000000803 UP000192221 UP000092445 UP000092443 UP000092460 UP000002282 UP000008711 UP000091820 UP000001070 UP000001292
PRIDE
Pfam
Interpro
IPR032503
FAO_M
+ More
IPR027266 TrmE/GcvT_dom1
IPR006222 GCV_T_N
IPR036188 FAD/NAD-bd_sf
IPR029043 GcvT/YgfZ_C
IPR013977 GCV_T_C
IPR006076 FAD-dep_OxRdtase
IPR001295 Dihydroorotate_DH_CS
IPR009051 Helical_ferredxn
IPR005720 Dihydroorotate_DH
IPR013785 Aldolase_TIM
IPR028261 DPD_II
IPR017896 4Fe4S_Fe-S-bd
IPR017900 4Fe4S_Fe_S_CS
IPR023753 FAD/NAD-binding_dom
IPR027266 TrmE/GcvT_dom1
IPR006222 GCV_T_N
IPR036188 FAD/NAD-bd_sf
IPR029043 GcvT/YgfZ_C
IPR013977 GCV_T_C
IPR006076 FAD-dep_OxRdtase
IPR001295 Dihydroorotate_DH_CS
IPR009051 Helical_ferredxn
IPR005720 Dihydroorotate_DH
IPR013785 Aldolase_TIM
IPR028261 DPD_II
IPR017896 4Fe4S_Fe-S-bd
IPR017900 4Fe4S_Fe_S_CS
IPR023753 FAD/NAD-binding_dom
ProteinModelPortal
A0A2W1BXT2
A0A194QU73
A0A212F9L9
A0A2J7QWV3
A0A2J7QWU2
D6WGA9
+ More
N6T053 E2C0J5 E2AFR4 A0A0J7L4H8 A0A1Q3FF35 A0A1Q3FFS8 A0A023F461 A0A182R2U8 A0A1Q3FFL9 A0A158NAM6 A0A1S4FMS9 A0A182WAU6 Q16VJ0 A0A182Q1A7 A0A023EWM8 A0A026WLF6 A0A224X726 A0A195DQN8 A0A195CLD2 A0A182NN92 A0A182IKB4 A0A151XIH2 Q7Q2M2 F4W6V4 A0A182LPU8 A0A182G1Y4 A0A067R5Z2 A0A182K281 A0A151I694 A0A2A3EJU0 A0A182HGJ4 A0A3L8DCT8 A0A182YI71 J9K0C5 B0XBZ6 A0A232EST7 A0A182PNG5 A0A084W1B7 A0A2M4CVA4 A0A2M4AI98 A0A2J7QWU4 A0A2S2QJN3 A0A0L7QZ50 A0A2H8TN07 A0A1B6D3W9 E9J3K1 U5EU87 A0A1B6FLK8 A0A154PB33 A0A2H1V324 A0A1B6KYV5 A0A1B6L0J7 E0VHC3 A0A1Y1K630 A0A0A9WV29 A0A146L4D4 T1PCJ5 A0A1I8M5W6 A0A182FLS3 A0A195EVI8 A0A1B0C9I4 W8B260 A0A034VKT0 A0A0K8U533 A0A1L8E2H1 A0A1I8PX73 B4M7A7 B4L5R6 A0A0L0BTF1 B3MXF7 A0A3B0KRM2 Q29J50 B4NP81 A0A0M4ETX9 A0A1A9VCK3 B4R4H6 A0A1B0GA17 Q9W4K8 A0A1W4V232 A0A1A9ZEI2 A0A1A9XBV0 A0A1B0BE01 B4Q063 B3NU25 A0A1A9WT20 Q8MQN6 B4JKC8 T1IHI3 B4I0Z6 A0A0P5RUW1 A0A0P6HC22 A0A0P6FBP3 A0A0P5S5F8
N6T053 E2C0J5 E2AFR4 A0A0J7L4H8 A0A1Q3FF35 A0A1Q3FFS8 A0A023F461 A0A182R2U8 A0A1Q3FFL9 A0A158NAM6 A0A1S4FMS9 A0A182WAU6 Q16VJ0 A0A182Q1A7 A0A023EWM8 A0A026WLF6 A0A224X726 A0A195DQN8 A0A195CLD2 A0A182NN92 A0A182IKB4 A0A151XIH2 Q7Q2M2 F4W6V4 A0A182LPU8 A0A182G1Y4 A0A067R5Z2 A0A182K281 A0A151I694 A0A2A3EJU0 A0A182HGJ4 A0A3L8DCT8 A0A182YI71 J9K0C5 B0XBZ6 A0A232EST7 A0A182PNG5 A0A084W1B7 A0A2M4CVA4 A0A2M4AI98 A0A2J7QWU4 A0A2S2QJN3 A0A0L7QZ50 A0A2H8TN07 A0A1B6D3W9 E9J3K1 U5EU87 A0A1B6FLK8 A0A154PB33 A0A2H1V324 A0A1B6KYV5 A0A1B6L0J7 E0VHC3 A0A1Y1K630 A0A0A9WV29 A0A146L4D4 T1PCJ5 A0A1I8M5W6 A0A182FLS3 A0A195EVI8 A0A1B0C9I4 W8B260 A0A034VKT0 A0A0K8U533 A0A1L8E2H1 A0A1I8PX73 B4M7A7 B4L5R6 A0A0L0BTF1 B3MXF7 A0A3B0KRM2 Q29J50 B4NP81 A0A0M4ETX9 A0A1A9VCK3 B4R4H6 A0A1B0GA17 Q9W4K8 A0A1W4V232 A0A1A9ZEI2 A0A1A9XBV0 A0A1B0BE01 B4Q063 B3NU25 A0A1A9WT20 Q8MQN6 B4JKC8 T1IHI3 B4I0Z6 A0A0P5RUW1 A0A0P6HC22 A0A0P6FBP3 A0A0P5S5F8
PDB
4PAB
E-value=3.74233e-93,
Score=875
Ontologies
GO
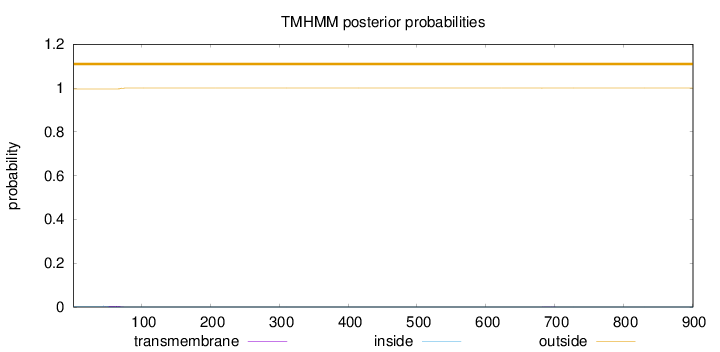
Topology
Length:
901
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0807700000000001
Exp number, first 60 AAs:
0.03841
Total prob of N-in:
0.00390
outside
1 - 901
Population Genetic Test Statistics
Pi
4.644179
Theta
16.387327
Tajima's D
-1.162696
CLR
0.056962
CSRT
0.104694765261737
Interpretation
Uncertain
