Gene
KWMTBOMO00083
Annotation
PREDICTED:_CUB_and_sushi_domain-containing_protein_3_isoform_X3_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.522
Sequence
CDS
ATGGATGTGCTGTACTTGATTGTCATCTTGCTGCAAGGCTATGTATCGGCAGCTTGTAACTTCCCGGGAGCGCCGGCCCATTGCCACGTGACGTTCTCCAACGACAGCCTGACAGAAGGCACCTCTGCCTGGTACGAATGTGATCGAGGCTTCGAACTGCTCGGTCCCTCTAGGAGGGTGTGTACGAGAGATGGAACGTGGATTCCTGAGGGAATACCGTTTTGCGGTGAGTACCATTATCGGGACTAA
Protein
MDVLYLIVILLQGYVSAACNFPGAPAHCHVTFSNDSLTEGTSAWYECDRGFELLGPSRRVCTRDGTWIPEGIPFCGEYHYRD
Summary
Uniprot
A0A2W1BC95
A0A0K2TB82
A0A0L7LTV7
A0A0K2TCA7
A0A194QUB1
T1J8K0
+ More
A0A1J1IWT4 A0A1A9WC40 A0A2R7W4C8 A0A212EQQ5 J9JN65 A0A1B0FJN9 S4PX17 T1HTS7 A0A2H8TVZ1 Q16GJ5 A0A1B0GNI7 A0A2S2P4M5 A0A194PTK1 A0A1D2N5Z5 B3NUU6 A8JUX3 W8ATQ6 A0A226DFR8 A0A1Y3AQH9 M9PHT4 Q9VXX7 B3MR68 X2JBY1 W5JTC7 A0A336K7D3 A0A1V1FQ92 B4R546 A0A087UCJ8 B5DKN4 A0A3B0JY53 B4IJA4 A0A1B0CWM9 A0A1V1FIP8 A0A1V1FKG6 A0A0A1X034 A0A034V5X2 A0A0K8UH90 A0A182YMX3 A0A0K8VP00 A0A0K8TYL6 B4JMK8 A0A182PUA4 A0A1I8MVF0 A0A1I8MVF2 B4L5B1 A0A1I8MVE9 A0A1I8MVF6 A0A1W4VFH6 A0A1W4V384 A0A0M3QZP2 A0A0P5SU90 A0A182RVU2 B4N223 A0A0N8E054 A0A0P5RDU4 A0A0P5N0P7 A0A0P6EIU2 A0A0P6EL03 A0A0P5VR32 A0A0P6JZT4 A0A0P6CRR8 A0A0N8BJS6 A0A0P5SYX8 A0A0P5DCC3 A0A0P5WAI6 A0A0N8B3M7 A0A0P5ED97 A0A0P5I4J9 A0A1I8PA81 A0A0P5JT14 A0A0N8B976 A0A0P5B5Z1 A0A1I8PA87 A0A0P5JX41 A0A1I8PAF5 A0A0P5DSH4 A0A0P5J907 A0A0N8C393 A0A0P5T4G0 B4MAA8 A0A0N7ZZ34 A0A0N7ZNK9 D6WGL3 E0VZZ7 A0A084VNV4 A0A182LRQ6 A0A0P5TWM3
A0A1J1IWT4 A0A1A9WC40 A0A2R7W4C8 A0A212EQQ5 J9JN65 A0A1B0FJN9 S4PX17 T1HTS7 A0A2H8TVZ1 Q16GJ5 A0A1B0GNI7 A0A2S2P4M5 A0A194PTK1 A0A1D2N5Z5 B3NUU6 A8JUX3 W8ATQ6 A0A226DFR8 A0A1Y3AQH9 M9PHT4 Q9VXX7 B3MR68 X2JBY1 W5JTC7 A0A336K7D3 A0A1V1FQ92 B4R546 A0A087UCJ8 B5DKN4 A0A3B0JY53 B4IJA4 A0A1B0CWM9 A0A1V1FIP8 A0A1V1FKG6 A0A0A1X034 A0A034V5X2 A0A0K8UH90 A0A182YMX3 A0A0K8VP00 A0A0K8TYL6 B4JMK8 A0A182PUA4 A0A1I8MVF0 A0A1I8MVF2 B4L5B1 A0A1I8MVE9 A0A1I8MVF6 A0A1W4VFH6 A0A1W4V384 A0A0M3QZP2 A0A0P5SU90 A0A182RVU2 B4N223 A0A0N8E054 A0A0P5RDU4 A0A0P5N0P7 A0A0P6EIU2 A0A0P6EL03 A0A0P5VR32 A0A0P6JZT4 A0A0P6CRR8 A0A0N8BJS6 A0A0P5SYX8 A0A0P5DCC3 A0A0P5WAI6 A0A0N8B3M7 A0A0P5ED97 A0A0P5I4J9 A0A1I8PA81 A0A0P5JT14 A0A0N8B976 A0A0P5B5Z1 A0A1I8PA87 A0A0P5JX41 A0A1I8PAF5 A0A0P5DSH4 A0A0P5J907 A0A0N8C393 A0A0P5T4G0 B4MAA8 A0A0N7ZZ34 A0A0N7ZNK9 D6WGL3 E0VZZ7 A0A084VNV4 A0A182LRQ6 A0A0P5TWM3
Pubmed
EMBL
KZ150173
PZC72598.1
HACA01005719
CDW23080.1
JTDY01000097
KOB78882.1
+ More
HACA01005720 CDW23081.1 KQ461108 KPJ09057.1 JH431955 CVRI01000061 CRL04020.1 KK854316 PTY14577.1 AGBW02013268 OWR43784.1 ABLF02040020 CCAG010020658 GAIX01005818 JAA86742.1 ACPB03003387 ACPB03003388 ACPB03003389 GFXV01006601 MBW18406.1 CH478269 EAT33365.1 AJVK01013467 AJVK01013468 GGMR01011237 MBY23856.1 KQ459593 KPI96741.1 LJIJ01000192 ODN00688.1 CH954180 EDV47044.1 AE014298 ABW09416.1 GAMC01014325 GAMC01014324 GAMC01014322 GAMC01014321 GAMC01014319 JAB92230.1 LNIX01000021 OXA43704.1 MUJZ01070503 OTF69435.1 AGB95404.1 BT004847 AAF48429.2 AAO45203.1 CH902622 EDV34273.2 AHN59735.1 ADMH02000140 ETN67627.1 UFQS01000005 UFQT01000005 SSW96910.1 SSX17297.1 FX985250 BAX07263.1 CM000366 EDX17947.1 KK119214 KFM75087.1 CH379063 EDY72084.1 OUUW01000003 SPP78689.1 CH480847 EDW51083.1 AJWK01032706 AJWK01032707 AJWK01032708 AJWK01032709 FX985248 BAX07261.1 FX985249 BAX07262.1 GBXI01010051 GBXI01007911 GBXI01004453 JAD04241.1 JAD06381.1 JAD09839.1 GAKP01020281 GAKP01020279 GAKP01020276 JAC38676.1 GDHF01026267 JAI26047.1 GDHF01011695 JAI40619.1 GDHF01032961 GDHF01015426 GDHF01010897 JAI19353.1 JAI36888.1 JAI41417.1 CH916371 EDV91951.1 CH933811 EDW06370.2 CP012528 ALC49763.1 GDIP01142538 JAL61176.1 CH963925 EDW78412.2 GDIQ01074836 JAN19901.1 GDIQ01102276 JAL49450.1 GDIQ01152919 JAK98806.1 GDIQ01265870 GDIQ01074837 JAN19900.1 GDIQ01074838 JAN19899.1 GDIP01096590 JAM07125.1 GDIQ01001529 JAN93208.1 GDIQ01088376 JAN06361.1 GDIQ01170851 JAK80874.1 GDIP01133473 JAL70241.1 GDIP01158171 JAJ65231.1 GDIP01090030 JAM13685.1 GDIQ01216043 JAK35682.1 GDIP01162172 JAJ61230.1 GDIQ01219448 JAK32277.1 GDIQ01199143 JAK52582.1 GDIQ01200451 JAK51274.1 GDIP01193573 JAJ29829.1 GDIQ01197796 JAK53929.1 GDIP01158172 JAJ65230.1 GDIQ01202948 JAK48777.1 GDIQ01119115 JAL32611.1 GDIP01133472 LRGB01000024 JAL70242.1 KZS21487.1 CH940655 EDW66167.1 GDIP01198465 JAJ24937.1 GDIP01227865 JAI95536.1 KQ971321 EFA01290.2 DS235854 EEB18953.1 ATLV01014929 ATLV01014930 KE524996 KFB39648.1 AXCM01003328 AXCM01003329 GDIP01120974 JAL82740.1
HACA01005720 CDW23081.1 KQ461108 KPJ09057.1 JH431955 CVRI01000061 CRL04020.1 KK854316 PTY14577.1 AGBW02013268 OWR43784.1 ABLF02040020 CCAG010020658 GAIX01005818 JAA86742.1 ACPB03003387 ACPB03003388 ACPB03003389 GFXV01006601 MBW18406.1 CH478269 EAT33365.1 AJVK01013467 AJVK01013468 GGMR01011237 MBY23856.1 KQ459593 KPI96741.1 LJIJ01000192 ODN00688.1 CH954180 EDV47044.1 AE014298 ABW09416.1 GAMC01014325 GAMC01014324 GAMC01014322 GAMC01014321 GAMC01014319 JAB92230.1 LNIX01000021 OXA43704.1 MUJZ01070503 OTF69435.1 AGB95404.1 BT004847 AAF48429.2 AAO45203.1 CH902622 EDV34273.2 AHN59735.1 ADMH02000140 ETN67627.1 UFQS01000005 UFQT01000005 SSW96910.1 SSX17297.1 FX985250 BAX07263.1 CM000366 EDX17947.1 KK119214 KFM75087.1 CH379063 EDY72084.1 OUUW01000003 SPP78689.1 CH480847 EDW51083.1 AJWK01032706 AJWK01032707 AJWK01032708 AJWK01032709 FX985248 BAX07261.1 FX985249 BAX07262.1 GBXI01010051 GBXI01007911 GBXI01004453 JAD04241.1 JAD06381.1 JAD09839.1 GAKP01020281 GAKP01020279 GAKP01020276 JAC38676.1 GDHF01026267 JAI26047.1 GDHF01011695 JAI40619.1 GDHF01032961 GDHF01015426 GDHF01010897 JAI19353.1 JAI36888.1 JAI41417.1 CH916371 EDV91951.1 CH933811 EDW06370.2 CP012528 ALC49763.1 GDIP01142538 JAL61176.1 CH963925 EDW78412.2 GDIQ01074836 JAN19901.1 GDIQ01102276 JAL49450.1 GDIQ01152919 JAK98806.1 GDIQ01265870 GDIQ01074837 JAN19900.1 GDIQ01074838 JAN19899.1 GDIP01096590 JAM07125.1 GDIQ01001529 JAN93208.1 GDIQ01088376 JAN06361.1 GDIQ01170851 JAK80874.1 GDIP01133473 JAL70241.1 GDIP01158171 JAJ65231.1 GDIP01090030 JAM13685.1 GDIQ01216043 JAK35682.1 GDIP01162172 JAJ61230.1 GDIQ01219448 JAK32277.1 GDIQ01199143 JAK52582.1 GDIQ01200451 JAK51274.1 GDIP01193573 JAJ29829.1 GDIQ01197796 JAK53929.1 GDIP01158172 JAJ65230.1 GDIQ01202948 JAK48777.1 GDIQ01119115 JAL32611.1 GDIP01133472 LRGB01000024 JAL70242.1 KZS21487.1 CH940655 EDW66167.1 GDIP01198465 JAJ24937.1 GDIP01227865 JAI95536.1 KQ971321 EFA01290.2 DS235854 EEB18953.1 ATLV01014929 ATLV01014930 KE524996 KFB39648.1 AXCM01003328 AXCM01003329 GDIP01120974 JAL82740.1
Proteomes
UP000037510
UP000053240
UP000183832
UP000091820
UP000007151
UP000007819
+ More
UP000092444 UP000015103 UP000008820 UP000092462 UP000053268 UP000094527 UP000008711 UP000000803 UP000198287 UP000007801 UP000000673 UP000000304 UP000054359 UP000001819 UP000268350 UP000001292 UP000092461 UP000076408 UP000001070 UP000075885 UP000095301 UP000009192 UP000192221 UP000092553 UP000075900 UP000007798 UP000095300 UP000076858 UP000008792 UP000007266 UP000009046 UP000030765 UP000075883
UP000092444 UP000015103 UP000008820 UP000092462 UP000053268 UP000094527 UP000008711 UP000000803 UP000198287 UP000007801 UP000000673 UP000000304 UP000054359 UP000001819 UP000268350 UP000001292 UP000092461 UP000076408 UP000001070 UP000075885 UP000095301 UP000009192 UP000192221 UP000092553 UP000075900 UP000007798 UP000095300 UP000076858 UP000008792 UP000007266 UP000009046 UP000030765 UP000075883
Interpro
IPR000436
Sushi_SCR_CCP_dom
+ More
IPR035976 Sushi/SCR/CCP_sf
IPR016187 CTDL_fold
IPR001304 C-type_lectin-like
IPR016186 C-type_lectin-like/link_sf
IPR008979 Galactose-bd-like_sf
IPR018378 C-type_lectin_CS
IPR006585 FTP1
IPR011009 Kinase-like_dom_sf
IPR000687 RIO_kinase
IPR018935 RIO_kinase_CS
IPR000421 FA58C
IPR035976 Sushi/SCR/CCP_sf
IPR016187 CTDL_fold
IPR001304 C-type_lectin-like
IPR016186 C-type_lectin-like/link_sf
IPR008979 Galactose-bd-like_sf
IPR018378 C-type_lectin_CS
IPR006585 FTP1
IPR011009 Kinase-like_dom_sf
IPR000687 RIO_kinase
IPR018935 RIO_kinase_CS
IPR000421 FA58C
Gene 3D
CDD
ProteinModelPortal
A0A2W1BC95
A0A0K2TB82
A0A0L7LTV7
A0A0K2TCA7
A0A194QUB1
T1J8K0
+ More
A0A1J1IWT4 A0A1A9WC40 A0A2R7W4C8 A0A212EQQ5 J9JN65 A0A1B0FJN9 S4PX17 T1HTS7 A0A2H8TVZ1 Q16GJ5 A0A1B0GNI7 A0A2S2P4M5 A0A194PTK1 A0A1D2N5Z5 B3NUU6 A8JUX3 W8ATQ6 A0A226DFR8 A0A1Y3AQH9 M9PHT4 Q9VXX7 B3MR68 X2JBY1 W5JTC7 A0A336K7D3 A0A1V1FQ92 B4R546 A0A087UCJ8 B5DKN4 A0A3B0JY53 B4IJA4 A0A1B0CWM9 A0A1V1FIP8 A0A1V1FKG6 A0A0A1X034 A0A034V5X2 A0A0K8UH90 A0A182YMX3 A0A0K8VP00 A0A0K8TYL6 B4JMK8 A0A182PUA4 A0A1I8MVF0 A0A1I8MVF2 B4L5B1 A0A1I8MVE9 A0A1I8MVF6 A0A1W4VFH6 A0A1W4V384 A0A0M3QZP2 A0A0P5SU90 A0A182RVU2 B4N223 A0A0N8E054 A0A0P5RDU4 A0A0P5N0P7 A0A0P6EIU2 A0A0P6EL03 A0A0P5VR32 A0A0P6JZT4 A0A0P6CRR8 A0A0N8BJS6 A0A0P5SYX8 A0A0P5DCC3 A0A0P5WAI6 A0A0N8B3M7 A0A0P5ED97 A0A0P5I4J9 A0A1I8PA81 A0A0P5JT14 A0A0N8B976 A0A0P5B5Z1 A0A1I8PA87 A0A0P5JX41 A0A1I8PAF5 A0A0P5DSH4 A0A0P5J907 A0A0N8C393 A0A0P5T4G0 B4MAA8 A0A0N7ZZ34 A0A0N7ZNK9 D6WGL3 E0VZZ7 A0A084VNV4 A0A182LRQ6 A0A0P5TWM3
A0A1J1IWT4 A0A1A9WC40 A0A2R7W4C8 A0A212EQQ5 J9JN65 A0A1B0FJN9 S4PX17 T1HTS7 A0A2H8TVZ1 Q16GJ5 A0A1B0GNI7 A0A2S2P4M5 A0A194PTK1 A0A1D2N5Z5 B3NUU6 A8JUX3 W8ATQ6 A0A226DFR8 A0A1Y3AQH9 M9PHT4 Q9VXX7 B3MR68 X2JBY1 W5JTC7 A0A336K7D3 A0A1V1FQ92 B4R546 A0A087UCJ8 B5DKN4 A0A3B0JY53 B4IJA4 A0A1B0CWM9 A0A1V1FIP8 A0A1V1FKG6 A0A0A1X034 A0A034V5X2 A0A0K8UH90 A0A182YMX3 A0A0K8VP00 A0A0K8TYL6 B4JMK8 A0A182PUA4 A0A1I8MVF0 A0A1I8MVF2 B4L5B1 A0A1I8MVE9 A0A1I8MVF6 A0A1W4VFH6 A0A1W4V384 A0A0M3QZP2 A0A0P5SU90 A0A182RVU2 B4N223 A0A0N8E054 A0A0P5RDU4 A0A0P5N0P7 A0A0P6EIU2 A0A0P6EL03 A0A0P5VR32 A0A0P6JZT4 A0A0P6CRR8 A0A0N8BJS6 A0A0P5SYX8 A0A0P5DCC3 A0A0P5WAI6 A0A0N8B3M7 A0A0P5ED97 A0A0P5I4J9 A0A1I8PA81 A0A0P5JT14 A0A0N8B976 A0A0P5B5Z1 A0A1I8PA87 A0A0P5JX41 A0A1I8PAF5 A0A0P5DSH4 A0A0P5J907 A0A0N8C393 A0A0P5T4G0 B4MAA8 A0A0N7ZZ34 A0A0N7ZNK9 D6WGL3 E0VZZ7 A0A084VNV4 A0A182LRQ6 A0A0P5TWM3
PDB
2GSX
E-value=0.0100921,
Score=84
Ontologies
Topology
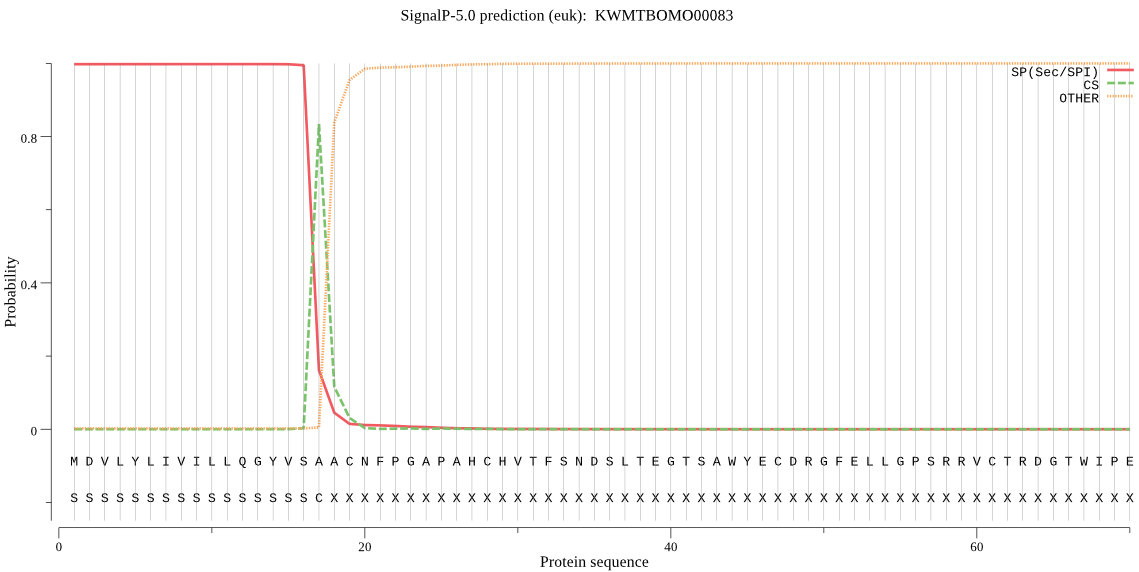
SignalP
Position: 1 - 17,
Likelihood: 0.997733
Length:
82
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.55763
Exp number, first 60 AAs:
4.55763
Total prob of N-in:
0.18275
outside
1 - 82

Population Genetic Test Statistics
Pi
26.041071
Theta
25.939546
Tajima's D
-1.049705
CLR
0.002236
CSRT
0.129143542822859
Interpretation
Uncertain
