Gene
KWMTBOMO00074
Pre Gene Modal
BGIBMGA012392
Annotation
endonuclease_and_reverse_transcriptase-like_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.611
Sequence
CDS
ATGAATGTGGAAGGATTTAGAGGAAGAGGTAGACCTAAGAAGAAATGGATGGATTGTGTGAAAGACGATATGGGTAGGAGGGGAGTGAGCGAAGAAATGGTATATGATAGAAGAGTATGGAAGGAGCAAACATGTTGCGTCGACCCCAGCAATGTCAGGGGCCTACATAGCAACCTTGACGCCGTACACCACCACCTTGAGACGGCGCAGCCTGCCCTGCTTTTTTTAACGGAGACGCAGATATCTGCTCCGGATGATACCTCATACCTTGAGTACCCCGGCTACGTATTGGAGCACAACTTCCTGCGTAAAGCCGGGGTGTGCGTATTCGTCCGGGCTGATGTCTGTTGTCGCCGTCTTCGGGGCCTCGAACAACGGGACCTGTCCCTCTTGTGGCTGCGCGTAGACCACGGGGGCCGTAGCCGAATCTACGCGTGCCTGTACAGGTCCCACAGCAGTGATGCAGGTTCAGCTCTGTTTGAGCATGTGCAAGAGGGGACTAACCGCGTGCTTGAGCAGTACCCATCTGCGGAGGTGGTGGTTCTTGGAGACTTTAACGCTCACCACCAAGAGTGGTTGGGGTCCAGAACCACTGACCTCCCGGGTCGGACTGCCTACGATTTCGCCTTGGCCTACGGCTTCTCCCAGCTGGTGACACAGCCCACCCGTGTCCCAGATATCGAGGGGCACGAGCCTTCTTTGTTGGACCTTCTGCTGACCACAGATCCGGCCGGATACAGTGTGGTGGTCGACGCTCCGCTTGGATCGTCTGATCACTGCCTTATTCGTGCTGCCATACCACTCTCTCGTCCTGATCGTCGAACGACGACCGGGTATCGAAGAGTTTGGCGGTATATGTCAGCAGATTGGGATGGATTGCGTGAATTTTACGCATCCTACCCATGGGGGCGGTTCTGCTTTTCCTCTGCTGATCCTGACGTCTGTGCGGACCGTCTTAAAGACGTGGTGCTCCAGGGGATGGAATTGTTTATTCCCTCCTCTGAAGTGCCCGTTGGGGGTCGCAGCAGACCCTGGTATAACAATGCCAGCAGGGATGCTGCACGCCTCAAGCGGTCCGCATACGTGGCATGGGATAATGCCAGGAGACGTCGGGATCCTAACATCTCAGGGGAAAGGCGGAAATATAACGCCGCTTCCAGGTCCTACAAGAAGGTCTAG
Protein
MNVEGFRGRGRPKKKWMDCVKDDMGRRGVSEEMVYDRRVWKEQTCCVDPSNVRGLHSNLDAVHHHLETAQPALLFLTETQISAPDDTSYLEYPGYVLEHNFLRKAGVCVFVRADVCCRRLRGLEQRDLSLLWLRVDHGGRSRIYACLYRSHSSDAGSALFEHVQEGTNRVLEQYPSAEVVVLGDFNAHHQEWLGSRTTDLPGRTAYDFALAYGFSQLVTQPTRVPDIEGHEPSLLDLLLTTDPAGYSVVVDAPLGSSDHCLIRAAIPLSRPDRRTTTGYRRVWRYMSADWDGLREFYASYPWGRFCFSSADPDVCADRLKDVVLQGMELFIPSSEVPVGGRSRPWYNNASRDAARLKRSAYVAWDNARRRRDPNISGERRKYNAASRSYKKV
Summary
Uniprot
Q6UV17
A0A2H1VKB9
A0A3D5S1K1
A0A147BMN4
A0A147BIC7
R7VLH1
+ More
D7ELX0 D6WYM1 A0A2H9T4N9 D7GYL9 A0A2H9T441 A0A2B4S280 A0A0P4VVV8 A0A147BJ61 A0A0P4VPK5 A0A1X7T6S7 A0A1E1XH11 W4XNU8 A0A131Y4V6 A0A2B4SC00 D6WYM2 A0A354GGU8 A0A147BM11 E3NN97 W4YYU9 A0A0A9Z9S6 A0A147BMC6 E3LJB3 A0A2P4VQ89 A0A147BQ97 A0A147BJ69 A0A147BJJ5 L7MD77 A0A0P4VXB7 A0A2G8L3E5 W4XMI9 L7LSG9 L7LSU8 A0A210QEX7 V4AQN4 E3MBS8 W4XIC7 A0A147BBB4 A0A147AJ39
D7ELX0 D6WYM1 A0A2H9T4N9 D7GYL9 A0A2H9T441 A0A2B4S280 A0A0P4VVV8 A0A147BJ61 A0A0P4VPK5 A0A1X7T6S7 A0A1E1XH11 W4XNU8 A0A131Y4V6 A0A2B4SC00 D6WYM2 A0A354GGU8 A0A147BM11 E3NN97 W4YYU9 A0A0A9Z9S6 A0A147BMC6 E3LJB3 A0A2P4VQ89 A0A147BQ97 A0A147BJ69 A0A147BJJ5 L7MD77 A0A0P4VXB7 A0A2G8L3E5 W4XMI9 L7LSG9 L7LSU8 A0A210QEX7 V4AQN4 E3MBS8 W4XIC7 A0A147BBB4 A0A147AJ39
Pubmed
EMBL
AY359886
AAQ57129.1
ODYU01003026
SOQ41278.1
DPOC01000269
HCX22453.1
+ More
GEGO01003370 JAR92034.1 GEGO01004865 JAR90539.1 AMQN01004026 AMQN01004027 KB292432 ELU17620.1 DS497938 EFA12433.1 KQ971357 EFA08424.1 NSIT01000234 PJE78200.1 GG695857 EFA13460.1 NSIT01000300 PJE77993.1 LSMT01000166 PFX24804.1 GDRN01111277 JAI56804.1 GEGO01004596 JAR90808.1 GDRN01111278 JAI56803.1 GFAC01000857 JAT98331.1 AAGJ04027320 GEFM01002248 JAP73548.1 LSMT01000104 PFX27391.1 EFA08423.1 DNUT01000248 HBI40236.1 GEGO01003567 JAR91837.1 DS269182 EFP10381.1 AAGJ04114595 GBHO01002973 JAG40631.1 GEGO01003481 JAR91923.1 DS268409 EFO94840.1 LFJK02000209 POM39446.1 GEGO01002473 JAR92931.1 GEGO01004630 JAR90774.1 GEGO01004441 JAR90963.1 GACK01003890 JAA61144.1 GDRN01110859 JAI56853.1 MRZV01000236 PIK54725.1 AAGJ04112043 GACK01010289 JAA54745.1 GACK01009888 JAA55146.1 NEDP02004020 OWF47171.1 KB201037 ESO99552.1 DS268433 EFO97776.1 AAGJ04072358 GEGO01007351 JAR88053.1 GCES01007896 JAR78427.1
GEGO01003370 JAR92034.1 GEGO01004865 JAR90539.1 AMQN01004026 AMQN01004027 KB292432 ELU17620.1 DS497938 EFA12433.1 KQ971357 EFA08424.1 NSIT01000234 PJE78200.1 GG695857 EFA13460.1 NSIT01000300 PJE77993.1 LSMT01000166 PFX24804.1 GDRN01111277 JAI56804.1 GEGO01004596 JAR90808.1 GDRN01111278 JAI56803.1 GFAC01000857 JAT98331.1 AAGJ04027320 GEFM01002248 JAP73548.1 LSMT01000104 PFX27391.1 EFA08423.1 DNUT01000248 HBI40236.1 GEGO01003567 JAR91837.1 DS269182 EFP10381.1 AAGJ04114595 GBHO01002973 JAG40631.1 GEGO01003481 JAR91923.1 DS268409 EFO94840.1 LFJK02000209 POM39446.1 GEGO01002473 JAR92931.1 GEGO01004630 JAR90774.1 GEGO01004441 JAR90963.1 GACK01003890 JAA61144.1 GDRN01110859 JAI56853.1 MRZV01000236 PIK54725.1 AAGJ04112043 GACK01010289 JAA54745.1 GACK01009888 JAA55146.1 NEDP02004020 OWF47171.1 KB201037 ESO99552.1 DS268433 EFO97776.1 AAGJ04072358 GEGO01007351 JAR88053.1 GCES01007896 JAR78427.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
Q6UV17
A0A2H1VKB9
A0A3D5S1K1
A0A147BMN4
A0A147BIC7
R7VLH1
+ More
D7ELX0 D6WYM1 A0A2H9T4N9 D7GYL9 A0A2H9T441 A0A2B4S280 A0A0P4VVV8 A0A147BJ61 A0A0P4VPK5 A0A1X7T6S7 A0A1E1XH11 W4XNU8 A0A131Y4V6 A0A2B4SC00 D6WYM2 A0A354GGU8 A0A147BM11 E3NN97 W4YYU9 A0A0A9Z9S6 A0A147BMC6 E3LJB3 A0A2P4VQ89 A0A147BQ97 A0A147BJ69 A0A147BJJ5 L7MD77 A0A0P4VXB7 A0A2G8L3E5 W4XMI9 L7LSG9 L7LSU8 A0A210QEX7 V4AQN4 E3MBS8 W4XIC7 A0A147BBB4 A0A147AJ39
D7ELX0 D6WYM1 A0A2H9T4N9 D7GYL9 A0A2H9T441 A0A2B4S280 A0A0P4VVV8 A0A147BJ61 A0A0P4VPK5 A0A1X7T6S7 A0A1E1XH11 W4XNU8 A0A131Y4V6 A0A2B4SC00 D6WYM2 A0A354GGU8 A0A147BM11 E3NN97 W4YYU9 A0A0A9Z9S6 A0A147BMC6 E3LJB3 A0A2P4VQ89 A0A147BQ97 A0A147BJ69 A0A147BJJ5 L7MD77 A0A0P4VXB7 A0A2G8L3E5 W4XMI9 L7LSG9 L7LSU8 A0A210QEX7 V4AQN4 E3MBS8 W4XIC7 A0A147BBB4 A0A147AJ39
Ontologies
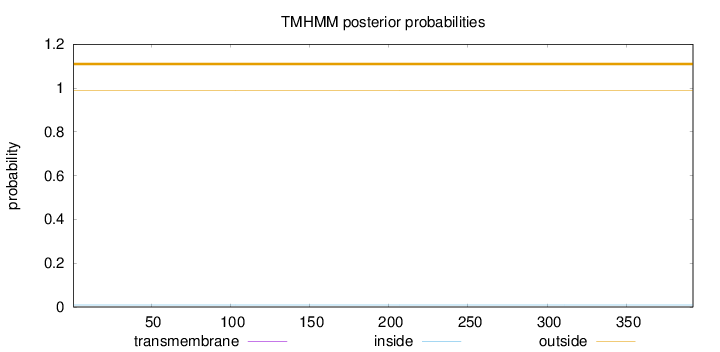
Topology
Length:
392
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00023
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00957
outside
1 - 392
Population Genetic Test Statistics
Pi
10.855065
Theta
15.075524
Tajima's D
-0.841501
CLR
0.311848
CSRT
0.166641667916604
Interpretation
Uncertain
