Pre Gene Modal
BGIBMGA002024
Annotation
PREDICTED:_plexin_domain-containing_protein_2_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.877 PlasmaMembrane Reliability : 1.507
Sequence
CDS
ATGGATAAAGCTATTGTGAATGTGAGATTTAAAGCAGCAGTGACTATTATTCTATTAGTATTCTTGGTCAACCGAGGCTCTCAAGCAGCTTGGCGCGAAGAATTCTTCACTCCGAGCTGGTTCTCTAAGAGTGCAACGACCGCAGCAGTGAGTCTCGATCGTCACCGTCGCGATGTTGCATCTTCGGCGAAGCCCGCGATTCCGGTCACGACCTCTGCAGTTCTTGTTACAAAGATGGACAAGATCACCAAGAATGACACCGAAATTGCCAATGTTACTGATGCTCTTCGGGCGGACGGCAACGTATCCGTGACTCAGATTCCACTTCTGGGCGTGAACGGCTCCGGCATCCTCAAGGTCACCCCGACCGAGAACACGCACAAAGTCAGCGAACCTATCGACCCTGACCCTGATTACGATATGCGAGACATTTCCAACTTTACCCCGGATAAAATAAAAGCGGAACACAACATAACCGATTTAACAATTGACAATCACGAGTTCTACAACAGCTCCTTCATCGGCAACGCTATGTTTTTCAACGAGTACTGGGCAAATATCACCACGAAGGGAGCTAATGCCCATCGCTTGCTCAGCAATTCCCACAGAAGGGCAACCATGATTCAACTGTCGTTCACATTCCCGTTTTACGGGTATCCAATCAAAGATATAACCGTGGCCACGGGAGGGTTCATTCACACCGGCGAGCACGTTCACAACTGGTTGGCCGCTACGCAATACATTGCTCCGTTGATGGCTAACTTCGACACTTCGCTCACCAACGACAGCTATGTTAAGCTTAAAGACGATGGCGACCGGTTCACGGTGCTCTGGGAGAACGTACCCCTCCAGGAGGACCTGACGAAGAAGTTCACGTTCGCGGCGACCCTATACAAGAACGGCGACATCGTGTTCGCCTACAAGAACATACCGATCGACGTCAACACAATAAAGAACACGAGCCATCCCGTCAAAGTGGGCATCAGCGACGCTTACATGACCGACGCGTTCTACTACCAGATCCCGAAGCTGACGATATACCACCGCGTCTCGTTCAAGAACCACAACATCAAGAACGGCACCGTCCTGTGGCTGACCGCGCTGCCGACCTGCATCCAGTTCAGCACGTGCGACGAGTGTCTCAACCACGACACCACGTTTAACTGTACGTGGTGCAGTCGTTTGCAGAAGTGCTCGAGCGGTACAGACAGGAACAAACAAAGCTGGGAGATCAAACAATGCAACGCGTCTGCACTGACGAACGCGACGACCTGCCCGGCGACGCAGTCCGACACTGCCAACAACACTGTCAACGCCAGCACCACAAATAAACCTGACAATCAATCAACGACCGCAGCCGATGCCGTTGATCACGCAAAATCTCGCGTTCAGATCTCAGTACCGGTTCAGAGGAGGCCGATAGGGGGCGCCGTGTTTGGGTTCGTGGTGGTCGCTCTGATATGTTCGCTTGCACTTTGGGTGGTGTACGCGTTCAAGAATCCTCACACTCGCAGCGGACAATTCCTTATTAAATACCGACCGTCGCAGTGGAGTTGGCGTCGCGGTGAAGCGCGCTACACGGCCGCCACCATCCACATGTAA
Protein
MDKAIVNVRFKAAVTIILLVFLVNRGSQAAWREEFFTPSWFSKSATTAAVSLDRHRRDVASSAKPAIPVTTSAVLVTKMDKITKNDTEIANVTDALRADGNVSVTQIPLLGVNGSGILKVTPTENTHKVSEPIDPDPDYDMRDISNFTPDKIKAEHNITDLTIDNHEFYNSSFIGNAMFFNEYWANITTKGANAHRLLSNSHRRATMIQLSFTFPFYGYPIKDITVATGGFIHTGEHVHNWLAATQYIAPLMANFDTSLTNDSYVKLKDDGDRFTVLWENVPLQEDLTKKFTFAATLYKNGDIVFAYKNIPIDVNTIKNTSHPVKVGISDAYMTDAFYYQIPKLTIYHRVSFKNHNIKNGTVLWLTALPTCIQFSTCDECLNHDTTFNCTWCSRLQKCSSGTDRNKQSWEIKQCNASALTNATTCPATQSDTANNTVNASTTNKPDNQSTTAADAVDHAKSRVQISVPVQRRPIGGAVFGFVVVALICSLALWVVYAFKNPHTRSGQFLIKYRPSQWSWRRGEARYTAATIHM
Summary
Uniprot
H9IXP1
A0A2W1BSG9
A0A194Q4R2
A0A212F9M9
A0A2H1V9V7
A0A1E1WPS9
+ More
A0A1L8EI55 A0A1L8EIB8 T1PLL2 A0A1A9VWY2 V5GJC2 A0A1I8MIH9 A0A1L8ECZ1 A0A1A9WT35 A0A034W0M1 A0A1I8PSQ6 W8BGN7 W8B4P7 A0A034VZ04 W8BS65 A0A1I8PSP2 A0A0M9A3B7 A0A0A1WNR2 A0A1I8PSL2 A0A034VVH0 B3MY22 W8B4P4 A0A2J7RRB2 A0A084W174 A0A034VY05 A0A067RE49 A0A088AIJ5 A0A1A9Z3V8 A0A2A3EJZ7 A0A1L8E3E4 A0A1L8E407 A0A1L8E3G3 A0A1L8E3E7 D6WNK8 A0A232F748 A0A1L8E3F3 A0A026WTW7 A0A0L7RD37 A0A1A9YUD4 B0WBH8 U5ERP6 A0A1S4FS14 Q16R67 A0A1L8E3F4 A0A3L8DA20 B4MJB0 A0A1B6DVW1 A0A1B6C4U2 E1ZZS4 A0A151JSS7 F4W7D1 A0A224XK12 B4R7J2 B4PXI1 A0A182Q8E6 B4IDP8 A0A151X8T1 A0A182YPR5 A0A195DSL0 A0A0Q9WNL8 A0A195ATE4 B3NW39 A0A146LRN5 A0A0K8S702 A0A158NPW0 A0A0A9Z303 A0A182NQ96 A0A1Q3F587 A0A1W4XPM5 A0A0R1EHW5 E2BAU8 A0A1B6G7J8 T1IEN7 Q9W2V9 K7J6U5 A0A0Q5T382 A0A1Q3F5A6 B4MEE4 A0A1Q3F5L9 A0A182RJV5 A0A336ML95 A0A1W6EW90 A0A1Y1MAB1 A0A0Q9W8W0 A0A2R7WXN0 A0A2M4BK79 A0A2M4BHV2 A0A0P5P5F3 A0A0P5H8T2 A0A0N8C6Z0 A0A0P5RRD6 A0A0P5P9M1 A0A0P5HZM8 A0A0P5WB76
A0A1L8EI55 A0A1L8EIB8 T1PLL2 A0A1A9VWY2 V5GJC2 A0A1I8MIH9 A0A1L8ECZ1 A0A1A9WT35 A0A034W0M1 A0A1I8PSQ6 W8BGN7 W8B4P7 A0A034VZ04 W8BS65 A0A1I8PSP2 A0A0M9A3B7 A0A0A1WNR2 A0A1I8PSL2 A0A034VVH0 B3MY22 W8B4P4 A0A2J7RRB2 A0A084W174 A0A034VY05 A0A067RE49 A0A088AIJ5 A0A1A9Z3V8 A0A2A3EJZ7 A0A1L8E3E4 A0A1L8E407 A0A1L8E3G3 A0A1L8E3E7 D6WNK8 A0A232F748 A0A1L8E3F3 A0A026WTW7 A0A0L7RD37 A0A1A9YUD4 B0WBH8 U5ERP6 A0A1S4FS14 Q16R67 A0A1L8E3F4 A0A3L8DA20 B4MJB0 A0A1B6DVW1 A0A1B6C4U2 E1ZZS4 A0A151JSS7 F4W7D1 A0A224XK12 B4R7J2 B4PXI1 A0A182Q8E6 B4IDP8 A0A151X8T1 A0A182YPR5 A0A195DSL0 A0A0Q9WNL8 A0A195ATE4 B3NW39 A0A146LRN5 A0A0K8S702 A0A158NPW0 A0A0A9Z303 A0A182NQ96 A0A1Q3F587 A0A1W4XPM5 A0A0R1EHW5 E2BAU8 A0A1B6G7J8 T1IEN7 Q9W2V9 K7J6U5 A0A0Q5T382 A0A1Q3F5A6 B4MEE4 A0A1Q3F5L9 A0A182RJV5 A0A336ML95 A0A1W6EW90 A0A1Y1MAB1 A0A0Q9W8W0 A0A2R7WXN0 A0A2M4BK79 A0A2M4BHV2 A0A0P5P5F3 A0A0P5H8T2 A0A0N8C6Z0 A0A0P5RRD6 A0A0P5P9M1 A0A0P5HZM8 A0A0P5WB76
Pubmed
19121390
28756777
26354079
22118469
25315136
25348373
+ More
24495485 25830018 17994087 24438588 24845553 18362917 19820115 28648823 24508170 17510324 30249741 20798317 21719571 17550304 25244985 26823975 21347285 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 28004739
24495485 25830018 17994087 24438588 24845553 18362917 19820115 28648823 24508170 17510324 30249741 20798317 21719571 17550304 25244985 26823975 21347285 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 28004739
EMBL
BABH01015750
BABH01015751
KZ149952
PZC76594.1
KQ459463
KPJ00532.1
+ More
AGBW02009564 OWR50442.1 ODYU01001419 SOQ37576.1 GDQN01002213 JAT88841.1 GFDG01000465 JAV18334.1 GFDG01000466 JAV18333.1 KA649604 AFP64233.1 GALX01004277 JAB64189.1 GFDG01002285 JAV16514.1 GAKP01011639 JAC47313.1 GAMC01010427 JAB96128.1 GAMC01010428 GAMC01010425 JAB96130.1 GAKP01011640 JAC47312.1 GAMC01010429 GAMC01010426 GAMC01010424 JAB96126.1 KQ435774 KOX75093.1 GBXI01013898 GBXI01013603 JAD00394.1 JAD00689.1 GAKP01011641 JAC47311.1 CH902630 EDV38637.1 GAMC01010430 JAB96125.1 NEVH01000611 PNF43355.1 ATLV01019240 KE525266 KFB43968.1 GAKP01011638 JAC47314.1 KK852521 KDR22116.1 KZ288222 PBC32047.1 GFDF01000824 JAV13260.1 GFDF01000822 JAV13262.1 GFDF01000823 JAV13261.1 GFDF01000821 JAV13263.1 KQ971342 EFA03767.1 NNAY01000836 OXU26263.1 GFDF01000826 JAV13258.1 KK107105 EZA59505.1 KQ414614 KOC68877.1 CCAG010019883 DS231879 EDS42417.1 GANO01003644 JAB56227.1 CH477723 EAT36889.1 GFDF01000814 JAV13270.1 QOIP01000011 RLU16758.1 CH963738 EDW72199.1 GEDC01007470 GEDC01003112 JAS29828.1 JAS34186.1 GEDC01028791 JAS08507.1 GL435522 EFN73297.1 KQ982014 KYN30618.1 GL887844 EGI69804.1 GFTR01007636 JAW08790.1 CM000366 EDX17551.1 CM000162 EDX02932.1 AXCN02001888 CH480830 EDW45706.1 KQ982409 KYQ56739.1 KQ980487 KYN15905.1 KRF97514.1 KQ976745 KYM75493.1 CH954180 EDV46172.1 GDHC01015480 GDHC01009292 JAQ03149.1 JAQ09337.1 GBRD01017333 GBRD01017332 JAG48495.1 ADTU01022770 ADTU01022771 ADTU01022772 ADTU01022773 ADTU01022774 GBHO01007424 JAG36180.1 GFDL01012320 JAV22725.1 KRK07025.1 GL446860 EFN87176.1 GECZ01022162 GECZ01021903 GECZ01014336 GECZ01011398 JAS47607.1 JAS47866.1 JAS55433.1 JAS58371.1 ACPB03011507 AE014298 AY069641 BT083419 AAF46578.1 AAL39786.1 ACQ91093.1 ALI51145.1 ALI51146.1 AAZX01001299 AAZX01011167 AAZX01017108 KQS29858.1 GFDL01012294 JAV22751.1 CH940664 EDW62919.1 GFDL01012181 JAV22864.1 UFQS01001240 UFQT01001240 SSX09890.1 SSX29613.1 KY563582 ARK19991.1 GEZM01036341 JAV82769.1 KRF80854.1 KK855893 PTY24286.1 GGFJ01004291 MBW53432.1 GGFJ01003499 MBW52640.1 GDIQ01135128 JAL16598.1 GDIQ01231249 JAK20476.1 GDIQ01108739 JAL42987.1 GDIQ01097089 JAL54637.1 GDIQ01137840 JAL13886.1 GDIQ01243445 JAK08280.1 GDIP01088556 JAM15159.1
AGBW02009564 OWR50442.1 ODYU01001419 SOQ37576.1 GDQN01002213 JAT88841.1 GFDG01000465 JAV18334.1 GFDG01000466 JAV18333.1 KA649604 AFP64233.1 GALX01004277 JAB64189.1 GFDG01002285 JAV16514.1 GAKP01011639 JAC47313.1 GAMC01010427 JAB96128.1 GAMC01010428 GAMC01010425 JAB96130.1 GAKP01011640 JAC47312.1 GAMC01010429 GAMC01010426 GAMC01010424 JAB96126.1 KQ435774 KOX75093.1 GBXI01013898 GBXI01013603 JAD00394.1 JAD00689.1 GAKP01011641 JAC47311.1 CH902630 EDV38637.1 GAMC01010430 JAB96125.1 NEVH01000611 PNF43355.1 ATLV01019240 KE525266 KFB43968.1 GAKP01011638 JAC47314.1 KK852521 KDR22116.1 KZ288222 PBC32047.1 GFDF01000824 JAV13260.1 GFDF01000822 JAV13262.1 GFDF01000823 JAV13261.1 GFDF01000821 JAV13263.1 KQ971342 EFA03767.1 NNAY01000836 OXU26263.1 GFDF01000826 JAV13258.1 KK107105 EZA59505.1 KQ414614 KOC68877.1 CCAG010019883 DS231879 EDS42417.1 GANO01003644 JAB56227.1 CH477723 EAT36889.1 GFDF01000814 JAV13270.1 QOIP01000011 RLU16758.1 CH963738 EDW72199.1 GEDC01007470 GEDC01003112 JAS29828.1 JAS34186.1 GEDC01028791 JAS08507.1 GL435522 EFN73297.1 KQ982014 KYN30618.1 GL887844 EGI69804.1 GFTR01007636 JAW08790.1 CM000366 EDX17551.1 CM000162 EDX02932.1 AXCN02001888 CH480830 EDW45706.1 KQ982409 KYQ56739.1 KQ980487 KYN15905.1 KRF97514.1 KQ976745 KYM75493.1 CH954180 EDV46172.1 GDHC01015480 GDHC01009292 JAQ03149.1 JAQ09337.1 GBRD01017333 GBRD01017332 JAG48495.1 ADTU01022770 ADTU01022771 ADTU01022772 ADTU01022773 ADTU01022774 GBHO01007424 JAG36180.1 GFDL01012320 JAV22725.1 KRK07025.1 GL446860 EFN87176.1 GECZ01022162 GECZ01021903 GECZ01014336 GECZ01011398 JAS47607.1 JAS47866.1 JAS55433.1 JAS58371.1 ACPB03011507 AE014298 AY069641 BT083419 AAF46578.1 AAL39786.1 ACQ91093.1 ALI51145.1 ALI51146.1 AAZX01001299 AAZX01011167 AAZX01017108 KQS29858.1 GFDL01012294 JAV22751.1 CH940664 EDW62919.1 GFDL01012181 JAV22864.1 UFQS01001240 UFQT01001240 SSX09890.1 SSX29613.1 KY563582 ARK19991.1 GEZM01036341 JAV82769.1 KRF80854.1 KK855893 PTY24286.1 GGFJ01004291 MBW53432.1 GGFJ01003499 MBW52640.1 GDIQ01135128 JAL16598.1 GDIQ01231249 JAK20476.1 GDIQ01108739 JAL42987.1 GDIQ01097089 JAL54637.1 GDIQ01137840 JAL13886.1 GDIQ01243445 JAK08280.1 GDIP01088556 JAM15159.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000078200
UP000095301
UP000091820
+ More
UP000095300 UP000053105 UP000007801 UP000235965 UP000030765 UP000027135 UP000005203 UP000092445 UP000242457 UP000007266 UP000215335 UP000053097 UP000053825 UP000092444 UP000002320 UP000008820 UP000279307 UP000007798 UP000000311 UP000078541 UP000007755 UP000000304 UP000002282 UP000075886 UP000001292 UP000075809 UP000076408 UP000078492 UP000078540 UP000008711 UP000005205 UP000075884 UP000192223 UP000008237 UP000015103 UP000000803 UP000002358 UP000008792 UP000075900
UP000095300 UP000053105 UP000007801 UP000235965 UP000030765 UP000027135 UP000005203 UP000092445 UP000242457 UP000007266 UP000215335 UP000053097 UP000053825 UP000092444 UP000002320 UP000008820 UP000279307 UP000007798 UP000000311 UP000078541 UP000007755 UP000000304 UP000002282 UP000075886 UP000001292 UP000075809 UP000076408 UP000078492 UP000078540 UP000008711 UP000005205 UP000075884 UP000192223 UP000008237 UP000015103 UP000000803 UP000002358 UP000008792 UP000075900
PRIDE
Interpro
SUPFAM
SSF56366
SSF56366
Gene 3D
ProteinModelPortal
H9IXP1
A0A2W1BSG9
A0A194Q4R2
A0A212F9M9
A0A2H1V9V7
A0A1E1WPS9
+ More
A0A1L8EI55 A0A1L8EIB8 T1PLL2 A0A1A9VWY2 V5GJC2 A0A1I8MIH9 A0A1L8ECZ1 A0A1A9WT35 A0A034W0M1 A0A1I8PSQ6 W8BGN7 W8B4P7 A0A034VZ04 W8BS65 A0A1I8PSP2 A0A0M9A3B7 A0A0A1WNR2 A0A1I8PSL2 A0A034VVH0 B3MY22 W8B4P4 A0A2J7RRB2 A0A084W174 A0A034VY05 A0A067RE49 A0A088AIJ5 A0A1A9Z3V8 A0A2A3EJZ7 A0A1L8E3E4 A0A1L8E407 A0A1L8E3G3 A0A1L8E3E7 D6WNK8 A0A232F748 A0A1L8E3F3 A0A026WTW7 A0A0L7RD37 A0A1A9YUD4 B0WBH8 U5ERP6 A0A1S4FS14 Q16R67 A0A1L8E3F4 A0A3L8DA20 B4MJB0 A0A1B6DVW1 A0A1B6C4U2 E1ZZS4 A0A151JSS7 F4W7D1 A0A224XK12 B4R7J2 B4PXI1 A0A182Q8E6 B4IDP8 A0A151X8T1 A0A182YPR5 A0A195DSL0 A0A0Q9WNL8 A0A195ATE4 B3NW39 A0A146LRN5 A0A0K8S702 A0A158NPW0 A0A0A9Z303 A0A182NQ96 A0A1Q3F587 A0A1W4XPM5 A0A0R1EHW5 E2BAU8 A0A1B6G7J8 T1IEN7 Q9W2V9 K7J6U5 A0A0Q5T382 A0A1Q3F5A6 B4MEE4 A0A1Q3F5L9 A0A182RJV5 A0A336ML95 A0A1W6EW90 A0A1Y1MAB1 A0A0Q9W8W0 A0A2R7WXN0 A0A2M4BK79 A0A2M4BHV2 A0A0P5P5F3 A0A0P5H8T2 A0A0N8C6Z0 A0A0P5RRD6 A0A0P5P9M1 A0A0P5HZM8 A0A0P5WB76
A0A1L8EI55 A0A1L8EIB8 T1PLL2 A0A1A9VWY2 V5GJC2 A0A1I8MIH9 A0A1L8ECZ1 A0A1A9WT35 A0A034W0M1 A0A1I8PSQ6 W8BGN7 W8B4P7 A0A034VZ04 W8BS65 A0A1I8PSP2 A0A0M9A3B7 A0A0A1WNR2 A0A1I8PSL2 A0A034VVH0 B3MY22 W8B4P4 A0A2J7RRB2 A0A084W174 A0A034VY05 A0A067RE49 A0A088AIJ5 A0A1A9Z3V8 A0A2A3EJZ7 A0A1L8E3E4 A0A1L8E407 A0A1L8E3G3 A0A1L8E3E7 D6WNK8 A0A232F748 A0A1L8E3F3 A0A026WTW7 A0A0L7RD37 A0A1A9YUD4 B0WBH8 U5ERP6 A0A1S4FS14 Q16R67 A0A1L8E3F4 A0A3L8DA20 B4MJB0 A0A1B6DVW1 A0A1B6C4U2 E1ZZS4 A0A151JSS7 F4W7D1 A0A224XK12 B4R7J2 B4PXI1 A0A182Q8E6 B4IDP8 A0A151X8T1 A0A182YPR5 A0A195DSL0 A0A0Q9WNL8 A0A195ATE4 B3NW39 A0A146LRN5 A0A0K8S702 A0A158NPW0 A0A0A9Z303 A0A182NQ96 A0A1Q3F587 A0A1W4XPM5 A0A0R1EHW5 E2BAU8 A0A1B6G7J8 T1IEN7 Q9W2V9 K7J6U5 A0A0Q5T382 A0A1Q3F5A6 B4MEE4 A0A1Q3F5L9 A0A182RJV5 A0A336ML95 A0A1W6EW90 A0A1Y1MAB1 A0A0Q9W8W0 A0A2R7WXN0 A0A2M4BK79 A0A2M4BHV2 A0A0P5P5F3 A0A0P5H8T2 A0A0N8C6Z0 A0A0P5RRD6 A0A0P5P9M1 A0A0P5HZM8 A0A0P5WB76
PDB
3AL9
E-value=0.027917,
Score=89
Ontologies
GO
PANTHER
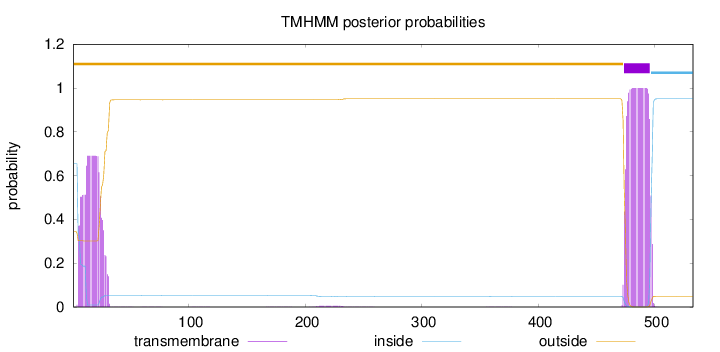
Topology
Subcellular location
Nucleus
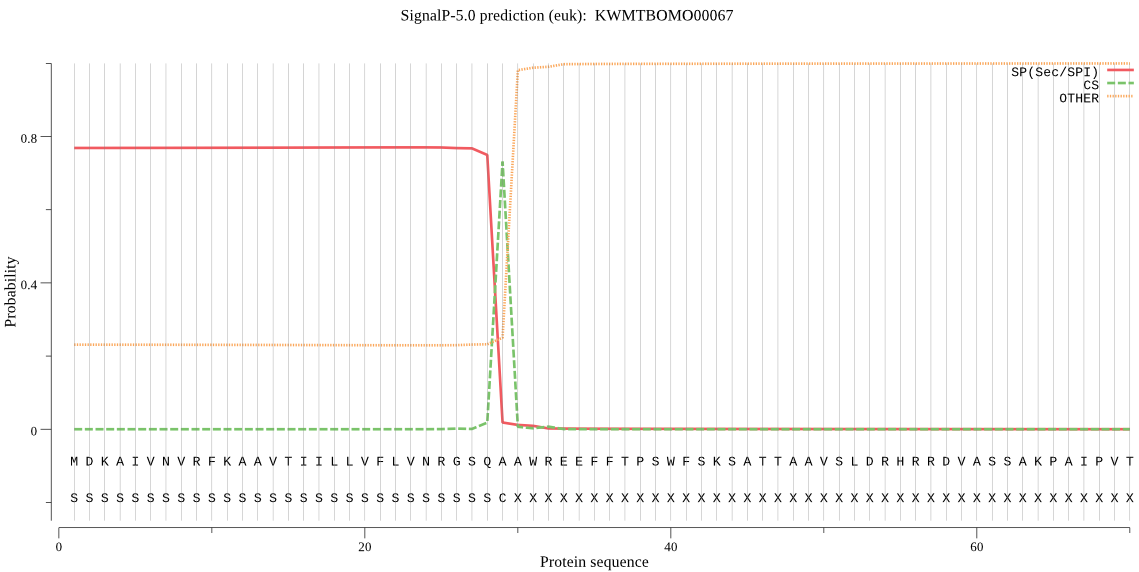
SignalP
Position: 1 - 29,
Likelihood: 0.769742
Length:
533
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
36.65779
Exp number, first 60 AAs:
13.88716
Total prob of N-in:
0.65402
POSSIBLE N-term signal
sequence
outside
1 - 473
TMhelix
474 - 496
inside
497 - 533
Population Genetic Test Statistics
Pi
18.638588
Theta
18.696369
Tajima's D
-1.248196
CLR
0.065026
CSRT
0.0919454027298635
Interpretation
Uncertain
