Gene
KWMTBOMO00065 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002099
Annotation
PREDICTED:_quinone_oxidoreductase_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.206
Sequence
CDS
ATGAGCAAATTGTGCAAACAAGTGTCGATCGAGTCCCCCGGACCGACCATAAAGGAATGTGTCTTTAGCTTTGACGTGCCAGTACCAGACGTGCCCCTCTACGGAGCTAGGATTAGAGTTGTGTACGCTGGAGCGTGTTACCGGGCTAACAGAACAAGATCGACTTCAGTGTGCAGTACTTCTTCTGTGTCTTCCATAGGCAGCTTTACTGGGGAGATGGATGGTTACGCTCATACCACGATTCCCGCCCATATCGGCATAAGAGACGGTGCGTTATTCCCTGGCTATGAAGTTGCGGGCATTATTGACGCCATAGGAAAGGCTGCCGAACGTACTGATGAACTCAAGGAAGGAGCAAGAATCGTTCTGTACCCTTACGAAGGGGTACCGCATGGTTATGCCGAATATATAGTGGTGCCCGACTTCAAATGTTTGGTGCCGATACCGGATCAATTGCCATTAAGCATAGCCGCTATGCTACCGACTGGCGCTTTGTTGGCCATGAATACAGTAGTATCGGCCAGTGAATGTTTAAAGGTGGCTTTAGAAGGACCTGCAAAGAAGGACAAAACGACTGTCCTAATTGTAGGAACTGGAGGCTTAGCCTTGTGGGCTCTACGCCTTGCCACGTACTATTTCAAGCGCGAAGAGTACAATGATCGCATTAGTATAGCTGTAGCTACGCTTAAGGATGAAGGATTTTTATTGGCGAAGGAGTGTGAAGAGGTCAACGTCGTGCAATGGAACGAAGATTTGTATGAGAAACAGTTGATAGAACGTACTACAGACGCGTGCGGTGGACCGGTCGACGTGGTACTCAACTTTGGCACCACTTCTCGATCTTTGCACCGTTCACTGCAGTGCTTGGCCCCTGAAGGTGTGGTCCTCGTGACCGAAGATGTAGGCGAGAGACTGCTGTCCAAGTTCGCGAAGAAGGCTGAAGATCTCAGCGTCAGTGTTGTGGTTGTGCCCAACGGGACCATCGAACAGCTGAAAGAACTGGTGTCACTAGTCGCACGCGGGGATATCGAACCCCCTCCGCACTCGGTGTACCCAGCTGAGGAGGCAGCAGAGGTCGTGCGAAAACTTTGTAATTCAGAAATAAAGGGACGAGCCATATTGAAGTTTCACAACGTGGAGTAA
Protein
MSKLCKQVSIESPGPTIKECVFSFDVPVPDVPLYGARIRVVYAGACYRANRTRSTSVCSTSSVSSIGSFTGEMDGYAHTTIPAHIGIRDGALFPGYEVAGIIDAIGKAAERTDELKEGARIVLYPYEGVPHGYAEYIVVPDFKCLVPIPDQLPLSIAAMLPTGALLAMNTVVSASECLKVALEGPAKKDKTTVLIVGTGGLALWALRLATYYFKREEYNDRISIAVATLKDEGFLLAKECEEVNVVQWNEDLYEKQLIERTTDACGGPVDVVLNFGTTSRSLHRSLQCLAPEGVVLVTEDVGERLLSKFAKKAEDLSVSVVVVPNGTIEQLKELVSLVARGDIEPPPHSVYPAEEAAEVVRKLCNSEIKGRAILKFHNVE
Summary
Uniprot
H9IXW6
A0A2W1BQK1
A0A2A4JC10
A0A2H1VLY5
A0A194QZP9
A0A194Q596
+ More
A0A212F9N1 A0A0L7LRN8 A0A2J7PPB4 A0A1Y1NDJ5 A0A1W4WKT4 A0A067RPX5 J3JXK6 Q17PH0 A0A1S4EVI9 A0A023EQK2 A0A182H639 V5GM40 Q7PVP3 A0A182PAL9 A0A182UAA7 A0A182XKT9 A0A1S4H0F4 A0A182I5A4 A0A2Y9D3E2 A0A1S4H0I3 A0A182KUA7 A0A158NIZ5 A0A226E7P4 A0A182WHA3 A0A182QT06 E2ANJ7 A0A182NF80 A0A182RG55 A0A2Y9D1U5 A0A182UP45 A0A1Q3FG57 A0A182Y9W3 A0A1Q3FGB0 U5ESH0 A0A1Q3FFZ1 A0A1Q3FGA3 A0A0J7L233 A0A182MWQ3 E9IIY5 B0W179 A0A026WE69 A0A336LTY0 A0A084VY97 A0A1D2M1R7 A0A195BBV1 A0A1A9VJ15 A0A1B0GBU7 A0A182IMR4 V9IDR2 A0A195DUE7 A0A195FHI6 A0A1A9Z7L7 K7IPW3 A0A2M3YZQ8 A0A2A3EUW5 V9IFH2 A0A2M4A2W0 A0A0R3NP48 A0A1I8PIY4 A0A1I8MGU0 A0A1I8PJ66 A0A2M4A279 T1PGP4 A0A1I8PJ53 A0A3L8E150 A0A1I8MGV0 A0A182F126 A0A2M4BPL9 A0A1I8MGU3 Q28YI9 T1DPK5 A0A3B0J355 A0A088ATI9 A0A0C9Q5I3 T1DEA3 A0A1A9X8P7 A0A1B0BYD5 A0A1A9XX75 W5JAY6 A0A0P8XPE5 A0A0K8U806 G3LFI2 A0A0K8UNF1 A0A0L7R7Z7 G3LFG9 B3MHB4 A0A0Q9XHR4 Q0E9G4 A0A0Q9W5I3 B4KRB5 B4HR32 B4QEC1 A0A0R1DTG2 A0A1W4V004
A0A212F9N1 A0A0L7LRN8 A0A2J7PPB4 A0A1Y1NDJ5 A0A1W4WKT4 A0A067RPX5 J3JXK6 Q17PH0 A0A1S4EVI9 A0A023EQK2 A0A182H639 V5GM40 Q7PVP3 A0A182PAL9 A0A182UAA7 A0A182XKT9 A0A1S4H0F4 A0A182I5A4 A0A2Y9D3E2 A0A1S4H0I3 A0A182KUA7 A0A158NIZ5 A0A226E7P4 A0A182WHA3 A0A182QT06 E2ANJ7 A0A182NF80 A0A182RG55 A0A2Y9D1U5 A0A182UP45 A0A1Q3FG57 A0A182Y9W3 A0A1Q3FGB0 U5ESH0 A0A1Q3FFZ1 A0A1Q3FGA3 A0A0J7L233 A0A182MWQ3 E9IIY5 B0W179 A0A026WE69 A0A336LTY0 A0A084VY97 A0A1D2M1R7 A0A195BBV1 A0A1A9VJ15 A0A1B0GBU7 A0A182IMR4 V9IDR2 A0A195DUE7 A0A195FHI6 A0A1A9Z7L7 K7IPW3 A0A2M3YZQ8 A0A2A3EUW5 V9IFH2 A0A2M4A2W0 A0A0R3NP48 A0A1I8PIY4 A0A1I8MGU0 A0A1I8PJ66 A0A2M4A279 T1PGP4 A0A1I8PJ53 A0A3L8E150 A0A1I8MGV0 A0A182F126 A0A2M4BPL9 A0A1I8MGU3 Q28YI9 T1DPK5 A0A3B0J355 A0A088ATI9 A0A0C9Q5I3 T1DEA3 A0A1A9X8P7 A0A1B0BYD5 A0A1A9XX75 W5JAY6 A0A0P8XPE5 A0A0K8U806 G3LFI2 A0A0K8UNF1 A0A0L7R7Z7 G3LFG9 B3MHB4 A0A0Q9XHR4 Q0E9G4 A0A0Q9W5I3 B4KRB5 B4HR32 B4QEC1 A0A0R1DTG2 A0A1W4V004
Pubmed
19121390
28756777
26354079
22118469
26227816
28004739
+ More
24845553 22516182 23537049 17510324 24945155 26483478 12364791 20966253 21347285 20798317 25244985 21282665 24508170 24438588 27289101 20075255 15632085 17994087 25315136 30249741 24330624 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304
24845553 22516182 23537049 17510324 24945155 26483478 12364791 20966253 21347285 20798317 25244985 21282665 24508170 24438588 27289101 20075255 15632085 17994087 25315136 30249741 24330624 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304
EMBL
BABH01015748
BABH01015749
KZ149952
PZC76591.1
NWSH01002025
PCG69389.1
+ More
ODYU01003279 SOQ41827.1 KQ461108 KPJ09066.1 KQ459463 KPJ00534.1 AGBW02009564 OWR50440.1 JTDY01000251 KOB78059.1 NEVH01023279 PNF18151.1 GEZM01005643 JAV96014.1 KK852539 KDR21784.1 APGK01039066 BT127975 KB740967 KB632399 AEE62937.1 ENN76789.1 ERL94724.1 CH477191 EAT48647.1 GAPW01001726 JAC11872.1 JXUM01113297 KQ565677 KXJ70789.1 GALX01003307 JAB65159.1 AAAB01008984 EAA43419.2 APCN01003842 ADTU01017255 ADTU01017256 LNIX01000006 OXA52891.1 AXCN02000825 GL441244 EFN64991.1 GFDL01008563 JAV26482.1 GFDL01008455 JAV26590.1 GANO01003246 JAB56625.1 GFDL01008579 JAV26466.1 GFDL01008474 JAV26571.1 LBMM01001243 KMQ96658.1 AXCM01005520 GL763562 EFZ19474.1 DS231820 EDS44053.1 KK107250 EZA54400.1 UFQS01000121 UFQT01000121 SSX00051.1 SSX20431.1 ATLV01018337 KE525231 KFB42941.1 LJIJ01006823 ODM86903.1 KQ976530 KYM81674.1 CCAG010005890 JR040643 AEY59220.1 KQ980334 KYN16503.1 KQ981523 KYN40160.1 GGFM01000970 MBW21721.1 KZ288185 PBC34939.1 JR040644 AEY59221.1 GGFK01001764 MBW35085.1 CM000071 KRT02714.1 GGFK01001530 MBW34851.1 KA647038 AFP61667.1 QOIP01000002 RLU25919.1 GGFJ01005885 MBW55026.1 EAL25976.2 GAMD01003269 JAA98321.1 OUUW01000001 SPP75715.1 GBYB01004091 GBYB01009323 JAG73858.1 JAG79090.1 GALA01001117 JAA93735.1 JXJN01022588 JXJN01022589 ADMH02001645 ETN61622.1 CH902619 KPU76490.1 GDHF01029603 JAI22711.1 JN374984 JN374987 AEO12081.1 AEO12084.1 GDHF01024107 JAI28207.1 KQ414637 KOC67007.1 JN374974 JN374988 AEO12071.1 AEO12085.1 EDV36891.1 CH933808 KRG04594.1 AE013599 JN374970 JN374971 JN374972 JN374973 JN374975 JN374976 JN374977 JN374978 JN374979 JN374980 JN374981 JN374982 JN374983 JN374985 JN374986 JN374989 JN374990 JN374991 JN374992 AAF59234.1 AAF59235.1 AEO12067.1 AEO12068.1 AEO12069.1 AEO12070.1 AEO12072.1 AEO12073.1 AEO12074.1 AEO12075.1 AEO12076.1 AEO12077.1 AEO12078.1 AEO12079.1 AEO12080.1 AEO12082.1 AEO12083.1 AEO12086.1 AEO12087.1 AEO12088.1 AEO12089.1 AGB93312.1 CH940648 KRF80300.1 EDW09331.1 CH480816 EDW46775.1 CM000362 CM002911 EDX06030.1 KMY92012.1 CM000157 KRJ98275.1
ODYU01003279 SOQ41827.1 KQ461108 KPJ09066.1 KQ459463 KPJ00534.1 AGBW02009564 OWR50440.1 JTDY01000251 KOB78059.1 NEVH01023279 PNF18151.1 GEZM01005643 JAV96014.1 KK852539 KDR21784.1 APGK01039066 BT127975 KB740967 KB632399 AEE62937.1 ENN76789.1 ERL94724.1 CH477191 EAT48647.1 GAPW01001726 JAC11872.1 JXUM01113297 KQ565677 KXJ70789.1 GALX01003307 JAB65159.1 AAAB01008984 EAA43419.2 APCN01003842 ADTU01017255 ADTU01017256 LNIX01000006 OXA52891.1 AXCN02000825 GL441244 EFN64991.1 GFDL01008563 JAV26482.1 GFDL01008455 JAV26590.1 GANO01003246 JAB56625.1 GFDL01008579 JAV26466.1 GFDL01008474 JAV26571.1 LBMM01001243 KMQ96658.1 AXCM01005520 GL763562 EFZ19474.1 DS231820 EDS44053.1 KK107250 EZA54400.1 UFQS01000121 UFQT01000121 SSX00051.1 SSX20431.1 ATLV01018337 KE525231 KFB42941.1 LJIJ01006823 ODM86903.1 KQ976530 KYM81674.1 CCAG010005890 JR040643 AEY59220.1 KQ980334 KYN16503.1 KQ981523 KYN40160.1 GGFM01000970 MBW21721.1 KZ288185 PBC34939.1 JR040644 AEY59221.1 GGFK01001764 MBW35085.1 CM000071 KRT02714.1 GGFK01001530 MBW34851.1 KA647038 AFP61667.1 QOIP01000002 RLU25919.1 GGFJ01005885 MBW55026.1 EAL25976.2 GAMD01003269 JAA98321.1 OUUW01000001 SPP75715.1 GBYB01004091 GBYB01009323 JAG73858.1 JAG79090.1 GALA01001117 JAA93735.1 JXJN01022588 JXJN01022589 ADMH02001645 ETN61622.1 CH902619 KPU76490.1 GDHF01029603 JAI22711.1 JN374984 JN374987 AEO12081.1 AEO12084.1 GDHF01024107 JAI28207.1 KQ414637 KOC67007.1 JN374974 JN374988 AEO12071.1 AEO12085.1 EDV36891.1 CH933808 KRG04594.1 AE013599 JN374970 JN374971 JN374972 JN374973 JN374975 JN374976 JN374977 JN374978 JN374979 JN374980 JN374981 JN374982 JN374983 JN374985 JN374986 JN374989 JN374990 JN374991 JN374992 AAF59234.1 AAF59235.1 AEO12067.1 AEO12068.1 AEO12069.1 AEO12070.1 AEO12072.1 AEO12073.1 AEO12074.1 AEO12075.1 AEO12076.1 AEO12077.1 AEO12078.1 AEO12079.1 AEO12080.1 AEO12082.1 AEO12083.1 AEO12086.1 AEO12087.1 AEO12088.1 AEO12089.1 AGB93312.1 CH940648 KRF80300.1 EDW09331.1 CH480816 EDW46775.1 CM000362 CM002911 EDX06030.1 KMY92012.1 CM000157 KRJ98275.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000235965 UP000192223 UP000027135 UP000019118 UP000030742 UP000008820 UP000069940 UP000249989 UP000007062 UP000075885 UP000075902 UP000076407 UP000075840 UP000075882 UP000005205 UP000198287 UP000075920 UP000075886 UP000000311 UP000075884 UP000075900 UP000075903 UP000076408 UP000036403 UP000075883 UP000002320 UP000053097 UP000030765 UP000094527 UP000078540 UP000078200 UP000092444 UP000075880 UP000078492 UP000078541 UP000092445 UP000002358 UP000242457 UP000001819 UP000095300 UP000095301 UP000279307 UP000069272 UP000268350 UP000005203 UP000092443 UP000092460 UP000000673 UP000007801 UP000053825 UP000009192 UP000000803 UP000008792 UP000001292 UP000000304 UP000002282 UP000192221
UP000235965 UP000192223 UP000027135 UP000019118 UP000030742 UP000008820 UP000069940 UP000249989 UP000007062 UP000075885 UP000075902 UP000076407 UP000075840 UP000075882 UP000005205 UP000198287 UP000075920 UP000075886 UP000000311 UP000075884 UP000075900 UP000075903 UP000076408 UP000036403 UP000075883 UP000002320 UP000053097 UP000030765 UP000094527 UP000078540 UP000078200 UP000092444 UP000075880 UP000078492 UP000078541 UP000092445 UP000002358 UP000242457 UP000001819 UP000095300 UP000095301 UP000279307 UP000069272 UP000268350 UP000005203 UP000092443 UP000092460 UP000000673 UP000007801 UP000053825 UP000009192 UP000000803 UP000008792 UP000001292 UP000000304 UP000002282 UP000192221
ProteinModelPortal
H9IXW6
A0A2W1BQK1
A0A2A4JC10
A0A2H1VLY5
A0A194QZP9
A0A194Q596
+ More
A0A212F9N1 A0A0L7LRN8 A0A2J7PPB4 A0A1Y1NDJ5 A0A1W4WKT4 A0A067RPX5 J3JXK6 Q17PH0 A0A1S4EVI9 A0A023EQK2 A0A182H639 V5GM40 Q7PVP3 A0A182PAL9 A0A182UAA7 A0A182XKT9 A0A1S4H0F4 A0A182I5A4 A0A2Y9D3E2 A0A1S4H0I3 A0A182KUA7 A0A158NIZ5 A0A226E7P4 A0A182WHA3 A0A182QT06 E2ANJ7 A0A182NF80 A0A182RG55 A0A2Y9D1U5 A0A182UP45 A0A1Q3FG57 A0A182Y9W3 A0A1Q3FGB0 U5ESH0 A0A1Q3FFZ1 A0A1Q3FGA3 A0A0J7L233 A0A182MWQ3 E9IIY5 B0W179 A0A026WE69 A0A336LTY0 A0A084VY97 A0A1D2M1R7 A0A195BBV1 A0A1A9VJ15 A0A1B0GBU7 A0A182IMR4 V9IDR2 A0A195DUE7 A0A195FHI6 A0A1A9Z7L7 K7IPW3 A0A2M3YZQ8 A0A2A3EUW5 V9IFH2 A0A2M4A2W0 A0A0R3NP48 A0A1I8PIY4 A0A1I8MGU0 A0A1I8PJ66 A0A2M4A279 T1PGP4 A0A1I8PJ53 A0A3L8E150 A0A1I8MGV0 A0A182F126 A0A2M4BPL9 A0A1I8MGU3 Q28YI9 T1DPK5 A0A3B0J355 A0A088ATI9 A0A0C9Q5I3 T1DEA3 A0A1A9X8P7 A0A1B0BYD5 A0A1A9XX75 W5JAY6 A0A0P8XPE5 A0A0K8U806 G3LFI2 A0A0K8UNF1 A0A0L7R7Z7 G3LFG9 B3MHB4 A0A0Q9XHR4 Q0E9G4 A0A0Q9W5I3 B4KRB5 B4HR32 B4QEC1 A0A0R1DTG2 A0A1W4V004
A0A212F9N1 A0A0L7LRN8 A0A2J7PPB4 A0A1Y1NDJ5 A0A1W4WKT4 A0A067RPX5 J3JXK6 Q17PH0 A0A1S4EVI9 A0A023EQK2 A0A182H639 V5GM40 Q7PVP3 A0A182PAL9 A0A182UAA7 A0A182XKT9 A0A1S4H0F4 A0A182I5A4 A0A2Y9D3E2 A0A1S4H0I3 A0A182KUA7 A0A158NIZ5 A0A226E7P4 A0A182WHA3 A0A182QT06 E2ANJ7 A0A182NF80 A0A182RG55 A0A2Y9D1U5 A0A182UP45 A0A1Q3FG57 A0A182Y9W3 A0A1Q3FGB0 U5ESH0 A0A1Q3FFZ1 A0A1Q3FGA3 A0A0J7L233 A0A182MWQ3 E9IIY5 B0W179 A0A026WE69 A0A336LTY0 A0A084VY97 A0A1D2M1R7 A0A195BBV1 A0A1A9VJ15 A0A1B0GBU7 A0A182IMR4 V9IDR2 A0A195DUE7 A0A195FHI6 A0A1A9Z7L7 K7IPW3 A0A2M3YZQ8 A0A2A3EUW5 V9IFH2 A0A2M4A2W0 A0A0R3NP48 A0A1I8PIY4 A0A1I8MGU0 A0A1I8PJ66 A0A2M4A279 T1PGP4 A0A1I8PJ53 A0A3L8E150 A0A1I8MGV0 A0A182F126 A0A2M4BPL9 A0A1I8MGU3 Q28YI9 T1DPK5 A0A3B0J355 A0A088ATI9 A0A0C9Q5I3 T1DEA3 A0A1A9X8P7 A0A1B0BYD5 A0A1A9XX75 W5JAY6 A0A0P8XPE5 A0A0K8U806 G3LFI2 A0A0K8UNF1 A0A0L7R7Z7 G3LFG9 B3MHB4 A0A0Q9XHR4 Q0E9G4 A0A0Q9W5I3 B4KRB5 B4HR32 B4QEC1 A0A0R1DTG2 A0A1W4V004
PDB
6FN6
E-value=0.000312587,
Score=104
Ontologies
GO
Topology
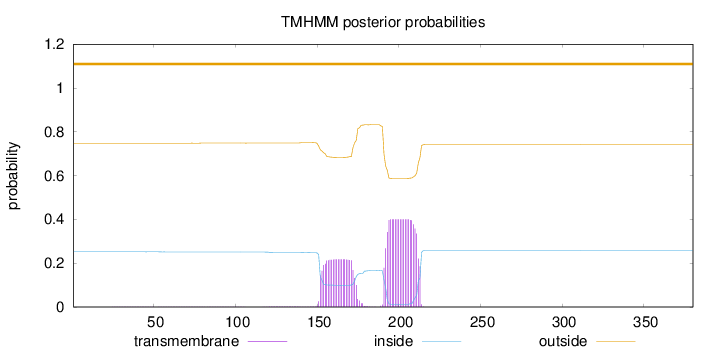
Length:
380
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
13.07271
Exp number, first 60 AAs:
0.01172
Total prob of N-in:
0.25193
outside
1 - 380
Population Genetic Test Statistics
Pi
18.298322
Theta
17.920163
Tajima's D
0.016072
CLR
0.022116
CSRT
0.372281385930703
Interpretation
Uncertain
