Gene
KWMTBOMO00062
Pre Gene Modal
BGIBMGA002957
Annotation
PREDICTED:_zinc_finger_CCHC_domain-containing_protein_13-like_[Bombyx_mori]
Full name
Gag polyprotein
+ More
Gag-Pol polyprotein
Gag-Pol polyprotein
Alternative Name
Pr160Gag-Pol
Location in the cell
Nuclear Reliability : 1.81
Sequence
CDS
ATGCGACGTGTCACCCAAACCGATGCGGAGGTTATTCCAACGTTCCGGCCGGATGGAAAATCTAGCAACGTCAAGGGATGGCTGCACAAGATCGACCAGTTGGATCACGTATATGGATGGGACAACAAAGACTGCCAGTTCATCATGCAGATATGTCTTCGTGGGTCGGCTAGGGATTGTGCGATACACCACCGCAACTATACTACACATCCGCGAATGGCTACCACATGGAGAAAAGAAGGTGCTGTTGCTCCAGTTGCAAGAGGGACCGACTTTCATCCAAAGAAATGTTACGCCTGCCGAAGAGAAGGTCATGAAACAAAGAACTGCAAAGAGCCGCGCTGCGAGGTATGCCATCGCCCGGGACACACGTCGGCCAGCTGA
Protein
MRRVTQTDAEVIPTFRPDGKSSNVKGWLHKIDQLDHVYGWDNKDCQFIMQICLRGSARDCAIHHRNYTTHPRMATTWRKEGAVAPVARGTDFHPKKCYACRREGHETKNCKEPRCEVCHRPGHTSAS
Summary
Description
Gag-Pol polyprotein: Mediates, with Gag polyprotein, the essential events in virion assembly, including binding the plasma membrane, making the protein-protein interactions necessary to create spherical particles, recruiting the viral Env proteins, and packaging the genomic RNA via direct interactions with the RNA packaging sequence (Psi). Gag-Pol polyprotein may regulate its own translation, by the binding genomic RNA in the 5'-UTR. At low concentration, the polyprotein would promote translation, whereas at high concentration, the polyprotein would encapsidate genomic RNA and then shut off translation.
Matrix protein p17: Targets the polyprotein to the plasma membrane via a multipartite membrane-binding signal, that includes its myristoylated N-terminus. Matrix protein is part of the pre-integration complex. Implicated in the release from host cell mediated by Vpu. Binds to RNA.
Capsid protein p24: Forms the conical core that encapsulates the genomic RNA-nucleocapsid complex in the virion. Most core are conical, with only 7% tubular. The core is constituted by capsid protein hexamer subunits. The core is disassembled soon after virion entry (By similarity). Host restriction factors such as TRIM5-alpha or TRIMCyp bind retroviral capsids and cause premature capsid disassembly, leading to blocks in reverse transcription. Capsid restriction by TRIM5 is one of the factors which restricts HIV-1 to the human species. Host PIN1 apparently facilitates the virion uncoating. On the other hand, interactions with PDZD8 or CYPA stabilize the capsid.
Nucleocapsid protein p7: Encapsulates and protects viral dimeric unspliced genomic RNA (gRNA). Binds these RNAs through its zinc fingers. Acts as a nucleic acid chaperone which is involved in rearangement of nucleic acid secondary structure during gRNA retrotranscription. Also facilitates template switch leading to recombination. As part of the polyprotein, participates in gRNA dimerization, packaging, tRNA incorporation and virion assembly.
Protease: Aspartyl protease that mediates proteolytic cleavages of Gag and Gag-Pol polyproteins during or shortly after the release of the virion from the plasma membrane. Cleavages take place as an ordered, step-wise cascade to yield mature proteins. This process is called maturation. Displays maximal activity during the budding process just prior to particle release from the cell. Also cleaves Nef and Vif, probably concomitantly with viral structural proteins on maturation of virus particles. Hydrolyzes host EIF4GI and PABP1 in order to shut off the capped cellular mRNA translation. The resulting inhibition of cellular protein synthesis serves to ensure maximal viral gene expression and to evade host immune response (By similarity).
Reverse transcriptase/ribonuclease H: Multifunctional enzyme that converts the viral RNA genome into dsDNA in the cytoplasm, shortly after virus entry into the cell. This enzyme displays a DNA polymerase activity that can copy either DNA or RNA templates, and a ribonuclease H (RNase H) activity that cleaves the RNA strand of RNA-DNA heteroduplexes in a partially processive 3' to 5' endonucleasic mode. Conversion of viral genomic RNA into dsDNA requires many steps. A tRNA(3)-Lys binds to the primer-binding site (PBS) situated at the 5'-end of the viral RNA. RT uses the 3' end of the tRNA primer to perform a short round of RNA-dependent minus-strand DNA synthesis. The reading proceeds through the U5 region and ends after the repeated (R) region which is present at both ends of viral RNA. The portion of the RNA-DNA heteroduplex is digested by the RNase H, resulting in a ssDNA product attached to the tRNA primer. This ssDNA/tRNA hybridizes with the identical R region situated at the 3' end of viral RNA. This template exchange, known as minus-strand DNA strong stop transfer, can be either intra- or intermolecular. RT uses the 3' end of this newly synthesized short ssDNA to perform the RNA-dependent minus-strand DNA synthesis of the whole template. RNase H digests the RNA template except for two polypurine tracts (PPTs) situated at the 5'-end and near the center of the genome. It is not clear if both polymerase and RNase H activities are simultaneous. RNase H probably can proceed both in a polymerase-dependent (RNA cut into small fragments by the same RT performing DNA synthesis) and a polymerase-independent mode (cleavage of remaining RNA fragments by free RTs). Secondly, RT performs DNA-directed plus-strand DNA synthesis using the PPTs that have not been removed by RNase H as primers. PPTs and tRNA primers are then removed by RNase H. The 3' and 5' ssDNA PBS regions hybridize to form a circular dsDNA intermediate. Strand displacement synthesis by RT to the PBS and PPT ends produces a blunt ended, linear dsDNA copy of the viral genome that includes long terminal repeats (LTRs) at both ends.
Integrase: Catalyzes viral DNA integration into the host chromosome, by performing a series of DNA cutting and joining reactions. This enzyme activity takes place after virion entry into a cell and reverse transcription of the RNA genome in dsDNA. The first step in the integration process is 3' processing. This step requires a complex comprising the viral genome, matrix protein, Vpr and integrase. This complex is called the pre-integration complex (PIC). The integrase protein removes 2 nucleotides from each 3' end of the viral DNA, leaving recessed CA OH's at the 3' ends. In the second step, the PIC enters cell nucleus. This process is mediated through integrase and Vpr proteins, and allows the virus to infect a non dividing cell. This ability to enter the nucleus is specific of lentiviruses, other retroviruses cannot and rely on cell division to access cell chromosomes. In the third step, termed strand transfer, the integrase protein joins the previously processed 3' ends to the 5' ends of strands of target cellular DNA at the site of integration. The 5'-ends are produced by integrase-catalyzed staggered cuts, 5 bp apart. A Y-shaped, gapped, recombination intermediate results, with the 5'-ends of the viral DNA strands and the 3' ends of target DNA strands remaining unjoined, flanking a gap of 5 bp. The last step is viral DNA integration into host chromosome. This involves host DNA repair synthesis in which the 5 bp gaps between the unjoined strands are filled in and then ligated. Since this process occurs at both cuts flanking the HIV genome, a 5 bp duplication of host DNA is produced at the ends of HIV-1 integration. Alternatively, Integrase may catalyze the excision of viral DNA just after strand transfer, this is termed disintegration.
Matrix protein p17: Targets the polyprotein to the plasma membrane via a multipartite membrane-binding signal, that includes its myristoylated N-terminus. Matrix protein is part of the pre-integration complex. Implicated in the release from host cell mediated by Vpu. Binds to RNA.
Capsid protein p24: Forms the conical core that encapsulates the genomic RNA-nucleocapsid complex in the virion. Most core are conical, with only 7% tubular. The core is constituted by capsid protein hexamer subunits. The core is disassembled soon after virion entry (By similarity). Host restriction factors such as TRIM5-alpha or TRIMCyp bind retroviral capsids and cause premature capsid disassembly, leading to blocks in reverse transcription. Capsid restriction by TRIM5 is one of the factors which restricts HIV-1 to the human species. Host PIN1 apparently facilitates the virion uncoating. On the other hand, interactions with PDZD8 or CYPA stabilize the capsid.
Nucleocapsid protein p7: Encapsulates and protects viral dimeric unspliced genomic RNA (gRNA). Binds these RNAs through its zinc fingers. Acts as a nucleic acid chaperone which is involved in rearangement of nucleic acid secondary structure during gRNA retrotranscription. Also facilitates template switch leading to recombination. As part of the polyprotein, participates in gRNA dimerization, packaging, tRNA incorporation and virion assembly.
Protease: Aspartyl protease that mediates proteolytic cleavages of Gag and Gag-Pol polyproteins during or shortly after the release of the virion from the plasma membrane. Cleavages take place as an ordered, step-wise cascade to yield mature proteins. This process is called maturation. Displays maximal activity during the budding process just prior to particle release from the cell. Also cleaves Nef and Vif, probably concomitantly with viral structural proteins on maturation of virus particles. Hydrolyzes host EIF4GI and PABP1 in order to shut off the capped cellular mRNA translation. The resulting inhibition of cellular protein synthesis serves to ensure maximal viral gene expression and to evade host immune response (By similarity).
Reverse transcriptase/ribonuclease H: Multifunctional enzyme that converts the viral RNA genome into dsDNA in the cytoplasm, shortly after virus entry into the cell. This enzyme displays a DNA polymerase activity that can copy either DNA or RNA templates, and a ribonuclease H (RNase H) activity that cleaves the RNA strand of RNA-DNA heteroduplexes in a partially processive 3' to 5' endonucleasic mode. Conversion of viral genomic RNA into dsDNA requires many steps. A tRNA(3)-Lys binds to the primer-binding site (PBS) situated at the 5'-end of the viral RNA. RT uses the 3' end of the tRNA primer to perform a short round of RNA-dependent minus-strand DNA synthesis. The reading proceeds through the U5 region and ends after the repeated (R) region which is present at both ends of viral RNA. The portion of the RNA-DNA heteroduplex is digested by the RNase H, resulting in a ssDNA product attached to the tRNA primer. This ssDNA/tRNA hybridizes with the identical R region situated at the 3' end of viral RNA. This template exchange, known as minus-strand DNA strong stop transfer, can be either intra- or intermolecular. RT uses the 3' end of this newly synthesized short ssDNA to perform the RNA-dependent minus-strand DNA synthesis of the whole template. RNase H digests the RNA template except for two polypurine tracts (PPTs) situated at the 5'-end and near the center of the genome. It is not clear if both polymerase and RNase H activities are simultaneous. RNase H probably can proceed both in a polymerase-dependent (RNA cut into small fragments by the same RT performing DNA synthesis) and a polymerase-independent mode (cleavage of remaining RNA fragments by free RTs). Secondly, RT performs DNA-directed plus-strand DNA synthesis using the PPTs that have not been removed by RNase H as primers. PPTs and tRNA primers are then removed by RNase H. The 3' and 5' ssDNA PBS regions hybridize to form a circular dsDNA intermediate. Strand displacement synthesis by RT to the PBS and PPT ends produces a blunt ended, linear dsDNA copy of the viral genome that includes long terminal repeats (LTRs) at both ends.
Integrase: Catalyzes viral DNA integration into the host chromosome, by performing a series of DNA cutting and joining reactions. This enzyme activity takes place after virion entry into a cell and reverse transcription of the RNA genome in dsDNA. The first step in the integration process is 3' processing. This step requires a complex comprising the viral genome, matrix protein, Vpr and integrase. This complex is called the pre-integration complex (PIC). The integrase protein removes 2 nucleotides from each 3' end of the viral DNA, leaving recessed CA OH's at the 3' ends. In the second step, the PIC enters cell nucleus. This process is mediated through integrase and Vpr proteins, and allows the virus to infect a non dividing cell. This ability to enter the nucleus is specific of lentiviruses, other retroviruses cannot and rely on cell division to access cell chromosomes. In the third step, termed strand transfer, the integrase protein joins the previously processed 3' ends to the 5' ends of strands of target cellular DNA at the site of integration. The 5'-ends are produced by integrase-catalyzed staggered cuts, 5 bp apart. A Y-shaped, gapped, recombination intermediate results, with the 5'-ends of the viral DNA strands and the 3' ends of target DNA strands remaining unjoined, flanking a gap of 5 bp. The last step is viral DNA integration into host chromosome. This involves host DNA repair synthesis in which the 5 bp gaps between the unjoined strands are filled in and then ligated. Since this process occurs at both cuts flanking the HIV genome, a 5 bp duplication of host DNA is produced at the ends of HIV-1 integration. Alternatively, Integrase may catalyze the excision of viral DNA just after strand transfer, this is termed disintegration.
Catalytic Activity
3'-end directed exonucleolytic cleavage of viral RNA-DNA hybrid.
Endohydrolysis of RNA in RNA/DNA hybrids. Three different cleavage modes: 1. sequence-specific internal cleavage of RNA. Human immunodeficiency virus type 1 and Moloney murine leukemia virus enzymes prefer to cleave the RNA strand one nucleotide away from the RNA-DNA junction. 2. RNA 5'-end directed cleavage 13-19 nucleotides from the RNA end. 3. DNA 3'-end directed cleavage 15-20 nucleotides away from the primer terminus.
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Specific for a P1 residue that is hydrophobic, and P1' variable, but often Pro.
Endohydrolysis of RNA in RNA/DNA hybrids. Three different cleavage modes: 1. sequence-specific internal cleavage of RNA. Human immunodeficiency virus type 1 and Moloney murine leukemia virus enzymes prefer to cleave the RNA strand one nucleotide away from the RNA-DNA junction. 2. RNA 5'-end directed cleavage 13-19 nucleotides from the RNA end. 3. DNA 3'-end directed cleavage 15-20 nucleotides away from the primer terminus.
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Specific for a P1 residue that is hydrophobic, and P1' variable, but often Pro.
Cofactor
Mg(2+)
Subunit
Matrix protein p17: Homotrimer; further assembles as hexamers of trimers (By similarity). Matrix protein p17: Interacts with gp41 (via C-terminus) (By similarity). Matrix protein p17: interacts with host CALM1; this interaction induces a conformational change in the Matrix protein, triggering exposure of the myristate group (By similarity). Matrix protein p17: interacts with host AP3D1; this interaction allows the polyprotein trafficking to multivesicular bodies during virus assembly (By similarity). Matrix protein p17: Part of the pre-integration complex (PIC) which is composed of viral genome, matrix protein, Vpr and integrase (By similarity). Capsid protein p24: Homodimer; the homodimer further multimerizes as homohexamers or homopentamers. Capsid protein p24: Interacts with human PPIA/CYPA (By similarity); This interaction stabilizes the capsid. Capsid protein p24: Interacts with human NUP153 (By similarity). Capsid protein p24: Interacts with host PDZD8; this interaction stabilizes the capsid (By similarity). Capsid protein p24: Interacts with monkey TRIM5; this interaction destabilizes the capsid (By similarity).Protease: Homodimer, whose active site consists of two apposed aspartic acid residues. Reverse transcriptase/ribonuclease H: Heterodimer of p66 RT and p51 RT (RT p66/p51). Heterodimerization of RT is essential for DNA polymerase activity. Despite the sequence identities, p66 RT and p51 RT have distinct folding. Integrase: Homodimer; possibly can form homotetramer. Integrase: Part of the pre-integration complex (PIC) which is composed of viral genome, matrix protein, Vpr and integrase. Integrase: Interacts with human SMARCB1/INI1 and human PSIP1/LEDGF isoform 1. Integrase: Interacts with human KPNA3; this interaction might play a role in nuclear import of the pre-integration complex (By similarity). Integrase: Interacts with human NUP153; this interaction might play a role in nuclear import of the pre-integration complex (By similarity).
Miscellaneous
Reverse transcriptase/ribonuclease H: Error-prone enzyme that lacks a proof-reading function. High mutations rate is a direct consequence of this characteristic. RT also displays frequent template switching leading to high recombination rate. Recombination mostly occurs between homologous regions of the two copackaged RNA genomes. If these two RNA molecules derive from different viral strains, reverse transcription will give rise to highly recombinated proviral DNAs.
HIV-1 lineages are divided in three main groups, M (for Major), O (for Outlier), and N (for New, or Non-M, Non-O). The vast majority of strains found worldwide belong to the group M. Group O seems to be endemic to and largely confined to Cameroon and neighboring countries in West Central Africa, where these viruses represent a small minority of HIV-1 strains. The group N is represented by a limited number of isolates from Cameroonian persons. The group M is further subdivided in 9 clades or subtypes (A to D, F to H, J and K).
Resistance to inhibitors associated with mutations are observed both in viral protease and in reverse transcriptase. Most of the time, single mutations confer only a modest reduction in drug susceptibility. Combination of several mutations is usually required to develop a high-level drug resistance. These mutations are predominantly found in clade B viruses and not in other genotypes. They are listed in the clade B representative isolate HXB2 (AC P04585).
HIV-1 lineages are divided in three main groups, M (for Major), O (for Outlier), and N (for New, or Non-M, Non-O). The vast majority of strains found worldwide belong to the group M. Group O seems to be endemic to and largely confined to Cameroon and neighboring countries in West Central Africa, where these viruses represent a small minority of HIV-1 strains. The group N is represented by a limited number of isolates from Cameroonian persons. The group M is further subdivided in 9 clades or subtypes (A to D, F to H, J and K).
Resistance to inhibitors associated with mutations are observed both in viral protease and in reverse transcriptase. Most of the time, single mutations confer only a modest reduction in drug susceptibility. Combination of several mutations is usually required to develop a high-level drug resistance. These mutations are predominantly found in clade B viruses and not in other genotypes. They are listed in the clade B representative isolate HXB2 (AC P04585).
Similarity
Belongs to the primate lentivirus group gag polyprotein family.
Keywords
3D-structure
Activation of host caspases by virus
AIDS
Aspartyl protease
Capsid protein
DNA integration
DNA recombination
DNA-binding
DNA-directed DNA polymerase
Endonuclease
Eukaryotic host gene expression shutoff by virus
Eukaryotic host translation shutoff by virus
Host cell membrane
Host cytoplasm
Host endosome
Host gene expression shutoff by virus
Host membrane
Host nucleus
Host-virus interaction
Hydrolase
Lipid-binding
Lipoprotein
Magnesium
Membrane
Metal-binding
Modulation of host cell apoptosis by virus
Multifunctional enzyme
Myristate
Nuclease
Nucleotidyltransferase
Phosphoprotein
Protease
Repeat
Ribosomal frameshifting
RNA-binding
RNA-directed DNA polymerase
Transferase
Viral genome integration
Viral nucleoprotein
Viral penetration into host nucleus
Viral release from host cell
Virion
Virion maturation
Virus entry into host cell
Zinc
Zinc-finger
Feature
chain Gag-Pol polyprotein
peptide Spacer peptide 1
peptide Spacer peptide 1
Uniprot
Pubmed
EMBL
BABH01010590
BABH01024735
BABH01024736
KQ972323
KXZ75922.1
KF859747
+ More
AHC30187.1 L11769 AAA44689.1 U42720 AAB47723.1 AF484513 AAN73754.1 AF484494 AAN73583.1 MH971628 AYE55483.1 AJ519489 CAD59562.1 GQ431214 ACV94279.1 AAN73755.1 EU110090 ABW86715.1 GQ431487 ACV94462.1 M22639 GU245717 ADD31665.1 AF484502 AAN73655.1
AHC30187.1 L11769 AAA44689.1 U42720 AAB47723.1 AF484513 AAN73754.1 AF484494 AAN73583.1 MH971628 AYE55483.1 AJ519489 CAD59562.1 GQ431214 ACV94279.1 AAN73755.1 EU110090 ABW86715.1 GQ431487 ACV94462.1 M22639 GU245717 ADD31665.1 AF484502 AAN73655.1
Proteomes
Pfam
Interpro
IPR036875
Znf_CCHC_sf
+ More
IPR001878 Znf_CCHC
IPR025398 DUF4371
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR014817 Gag_p6
IPR000071 Lentvrl_matrix_N
IPR010999 Retrovr_matrix
IPR012344 Matrix_HIV/RSV_N
IPR000721 Gag_p24
IPR008916 Retrov_capsid_C
IPR008919 Retrov_capsid_N
IPR010659 RVT_connect
IPR036862 Integrase_C_dom_sf_retrovir
IPR001969 Aspartic_peptidase_AS
IPR003308 Integrase_Zn-bd_dom_N
IPR001995 Peptidase_A2_cat
IPR010661 RVT_thumb
IPR002156 RNaseH_domain
IPR018061 Retropepsins
IPR001037 Integrase_C_retrovir
IPR034170 Retropepsin-like_cat_dom
IPR017856 Integrase-like_N
IPR001878 Znf_CCHC
IPR025398 DUF4371
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR014817 Gag_p6
IPR000071 Lentvrl_matrix_N
IPR010999 Retrovr_matrix
IPR012344 Matrix_HIV/RSV_N
IPR000721 Gag_p24
IPR008916 Retrov_capsid_C
IPR008919 Retrov_capsid_N
IPR010659 RVT_connect
IPR036862 Integrase_C_dom_sf_retrovir
IPR001969 Aspartic_peptidase_AS
IPR003308 Integrase_Zn-bd_dom_N
IPR001995 Peptidase_A2_cat
IPR010661 RVT_thumb
IPR002156 RNaseH_domain
IPR018061 Retropepsins
IPR001037 Integrase_C_retrovir
IPR034170 Retropepsin-like_cat_dom
IPR017856 Integrase-like_N
SUPFAM
ProteinModelPortal
PDB
5O2U
E-value=0.0124511,
Score=84
Ontologies
GO
GO:0008270
GO:0003676
GO:0015074
GO:0005634
GO:0005198
GO:0042025
GO:0019013
GO:0030430
GO:0039702
GO:0003723
GO:0016032
GO:0004533
GO:0039657
GO:0046718
GO:0004190
GO:0006310
GO:0003964
GO:0016020
GO:0075732
GO:0020002
GO:0003677
GO:0003887
GO:0044826
GO:0075713
GO:0004523
GO:0055036
GO:0072494
GO:0008289
GO:0039651
GO:0016791
GO:0006470
GO:0005524
GO:0048013
GO:0005003
GO:0005887
GO:0007169
GO:0016021
Topology
Subcellular location
Virion
Host nucleus
Host cytoplasm
Host cell membrane
Host endosome These locations are linked to virus assembly sites. The main location is the cell membrane, but under some circumstances, late endosomal compartments can serve as productive sites for virion assembly. With evidence from 8 publications.
Host multivesicular body These locations are linked to virus assembly sites. The main location is the cell membrane, but under some circumstances, late endosomal compartments can serve as productive sites for virion assembly. With evidence from 8 publications.
Virion membrane
Host nucleus
Host cytoplasm
Host cell membrane
Host endosome These locations are linked to virus assembly sites. The main location is the cell membrane, but under some circumstances, late endosomal compartments can serve as productive sites for virion assembly. With evidence from 8 publications.
Host multivesicular body These locations are linked to virus assembly sites. The main location is the cell membrane, but under some circumstances, late endosomal compartments can serve as productive sites for virion assembly. With evidence from 8 publications.
Virion membrane
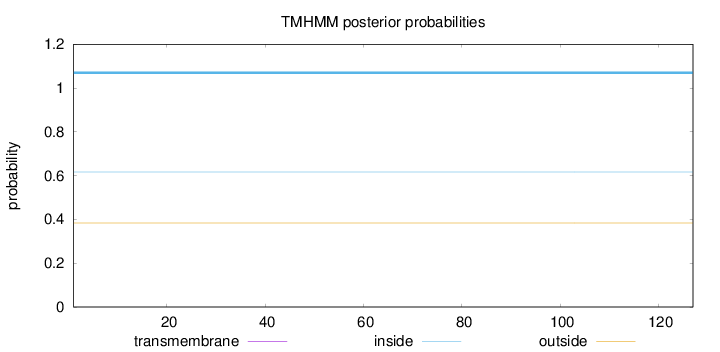
Length:
127
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.61674
inside
1 - 127
Population Genetic Test Statistics
Pi
5.606216
Theta
15.682873
Tajima's D
-1.822847
CLR
0
CSRT
0.0260486975651217
Interpretation
Uncertain
