Pre Gene Modal
BGIBMGA002096
Annotation
PREDICTED:_protein_tyrosine_phosphatase_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.137 Nuclear Reliability : 2.15
Sequence
CDS
ATGAATCCGAAGTTGAACAGCATTGAAGCCGAATATCTGGATATCATGCACAGGAATGGGTGGCCCCTGATTTATCAGAGGGTCGGTCAGGAGTGTCAAAAGCTCCCGTACACTTGTGTGGAGGCGAAGAGGCCTCACAACAAGGCGCTGAACCGCTACCGCGACGTGAACCCGTACGACCACAGCCGCATCGTCCTACAGCGTAGCGAGAACGATTACATAAACGCTAATCTCGTTAGGATGGAAAGCGCTGATCGCCAGTACATCCTGACTCAGGGACCGCTTGCGTTCACCGTAGGCCACTTTTGGCTGATGGTCTGGGAGCAGAACAGCCGCGCCGTCCTAATGCTGAACAAGGTCATCGAGAAAAACGAGATCAAGTGTCACTGGTACTGGCCCCATGGAAACGGCGAACAGCACAAGATGCTGCTAACAGATGTGAATCTTAGCGTCGAACAGATAAGCGAGGAGGAATGCCCAAACTACTCCATTCGGGTGCTTAAGCTATGCGATCTGGAGAGCTCCGAGAGTCGCGAGGTCATCCAGTTCCATTACACAACTTGGCCCGACTTCGGAGTGCCGTCTTCTCCTCACGCATTCCTCGAATACTTGAAGAAGATCCGCGATGCTGGCGCTTTGGAGAACGATGTTGGGCCAGCGGTCGTGCATTGCAGCGCCGGCATCGGTCGCTCTGGAACGTTCTGTCTCGTTGACTCCACTATGGTTATGATACAAAAGGCCGATGTGAAGTCTATCAACATCGAGCACATTATGCTTGAGATGCGCAAGTATCGCATGGGTCTAATTCAGACACCTGATCAGCTGCGTTTCTCGTACGAGGCCATCATCGAGGGGGCGAAGAGGGAGGACCCCAACTACGTACCTTCCAATATGGTCGAGAGCGTGTACTCGCCGGTGACTGACGTCGAAGATGATTGTCCTCCTCCGCCTCCTCCGCGCGGTGAGAGCCTCTGTCGCCCTCCTGCTCGTCCCTTGCCCGCGATTCCTTCAAGCATGTCTCTAGGCGAACTTCACCTGAGCGCTCCCGTCGAGTCTTCGAGCTCTGACGAATGTCCCCCCACTCCCCCAGAATTAGAACTACCGCCCTTCTCTTCAGCCGACGAAGATGAAGAACAGTCCGCCGTCCAGCCCGAGAGCAGCAGCGGCGCCAAGACGGAGACACTGCGCCGTCGTCGCATCCGCGAGCTGTCCGAGCGCGTCTCTCTGATGAAGCGACGGGCCCGCGACGCCGAACGATGGCACAACCTCAAGAGGTTTAAATTCGAAGAGAACAAAGAACATTGA
Protein
MNPKLNSIEAEYLDIMHRNGWPLIYQRVGQECQKLPYTCVEAKRPHNKALNRYRDVNPYDHSRIVLQRSENDYINANLVRMESADRQYILTQGPLAFTVGHFWLMVWEQNSRAVLMLNKVIEKNEIKCHWYWPHGNGEQHKMLLTDVNLSVEQISEEECPNYSIRVLKLCDLESSESREVIQFHYTTWPDFGVPSSPHAFLEYLKKIRDAGALENDVGPAVVHCSAGIGRSGTFCLVDSTMVMIQKADVKSINIEHIMLEMRKYRMGLIQTPDQLRFSYEAIIEGAKREDPNYVPSNMVESVYSPVTDVEDDCPPPPPPRGESLCRPPARPLPAIPSSMSLGELHLSAPVESSSSDECPPTPPELELPPFSSADEDEEQSAVQPESSSGAKTETLRRRRIRELSERVSLMKRRARDAERWHNLKRFKFEENKEH
Summary
Catalytic Activity
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
Uniprot
F2Z7K2
H9IXW3
A0A212F9N2
A0A2A4JEA4
A0A194Q4R7
A0A0M8ZR59
+ More
A0A194QVS8 A0A1Y1NGH0 A0A1Y1NLC4 V9IJC8 A0A2A3EMG7 A0A026WSH5 A0A088A9T3 A0A310S4Y0 A0A154P8J7 A0A0J7KY87 E2A118 A0A232FPK1 K7IW23 A0A1L8DLQ0 A0A1L8DLM4 A0A0A1XAE1 A0A1L8DM46 A0A182G3J1 A0A1S4F057 A0A0A1WR00 A0A1Q3F3X2 A0A139WLR4 A0A158NJ25 A0A023F0Y9 A0A1S4F055 A0A1Q3F3Z9 A0A195B6L2 D6WF82 A0A069DU32 A0A0L7QWY6 A0A1B6D5J2 A0A1A9W4A5 A0A151XK75 A0A1B6CAH5 E9IUJ4 A0A0V0G9N3 A0A0K8UPX5 A0A034VKG1 A0A0K8WC51 A0A182KW25 A0A224XD80 A0A1B0AVR3 A0A0L0BPA4 A0A034VLI5 F4WCH6 W8B1A4 A0A2M4BKY4 A0A195EL52 A0A0Q9XB75 B4KZ08 A0A2M4CPT6 A0A182LRF0 A0A0Q9WI44 A0A2M4BFJ4 B4LE80 A0A2M4AFC3 A0A2M4BFM3 A0A2M4AET3 A0A2W1BNF9 A0A195C5P4 A0A182R2L5 U5EYG1 B4J260 Q7PNX1 A0A1I8MBM0 A0A1I8MBM4 A0A182YNM0 A0A1I8NLY9 A0A1I8NLY3 A0A182XD23 A0A182HMF4 A0A182TTC4 A0A182WAL7
A0A194QVS8 A0A1Y1NGH0 A0A1Y1NLC4 V9IJC8 A0A2A3EMG7 A0A026WSH5 A0A088A9T3 A0A310S4Y0 A0A154P8J7 A0A0J7KY87 E2A118 A0A232FPK1 K7IW23 A0A1L8DLQ0 A0A1L8DLM4 A0A0A1XAE1 A0A1L8DM46 A0A182G3J1 A0A1S4F057 A0A0A1WR00 A0A1Q3F3X2 A0A139WLR4 A0A158NJ25 A0A023F0Y9 A0A1S4F055 A0A1Q3F3Z9 A0A195B6L2 D6WF82 A0A069DU32 A0A0L7QWY6 A0A1B6D5J2 A0A1A9W4A5 A0A151XK75 A0A1B6CAH5 E9IUJ4 A0A0V0G9N3 A0A0K8UPX5 A0A034VKG1 A0A0K8WC51 A0A182KW25 A0A224XD80 A0A1B0AVR3 A0A0L0BPA4 A0A034VLI5 F4WCH6 W8B1A4 A0A2M4BKY4 A0A195EL52 A0A0Q9XB75 B4KZ08 A0A2M4CPT6 A0A182LRF0 A0A0Q9WI44 A0A2M4BFJ4 B4LE80 A0A2M4AFC3 A0A2M4BFM3 A0A2M4AET3 A0A2W1BNF9 A0A195C5P4 A0A182R2L5 U5EYG1 B4J260 Q7PNX1 A0A1I8MBM0 A0A1I8MBM4 A0A182YNM0 A0A1I8NLY9 A0A1I8NLY3 A0A182XD23 A0A182HMF4 A0A182TTC4 A0A182WAL7
Pubmed
EMBL
AB543068
BAK19072.1
BABH01015741
BABH01015742
AGBW02009564
OWR50438.1
+ More
NWSH01001692 PCG70407.1 KQ459463 KPJ00537.1 KQ435891 KOX69445.1 KQ461108 KPJ09070.1 GEZM01003658 JAV96648.1 GEZM01003660 GEZM01003659 JAV96647.1 JR048067 AEY60571.1 KZ288212 PBC32880.1 KK107119 EZA58596.1 KQ773232 OAD52414.1 KQ434846 KZC08245.1 LBMM01002117 KMQ95253.1 GL435638 EFN72871.1 NNAY01000002 OXU32287.1 GFDF01006743 JAV07341.1 GFDF01006742 JAV07342.1 GBXI01006597 JAD07695.1 GFDF01006647 JAV07437.1 JXUM01040880 JXUM01040881 JXUM01040882 JXUM01040883 JXUM01040884 JXUM01040885 KQ561263 KXJ79163.1 GBXI01013464 GBXI01009827 JAD00828.1 JAD04465.1 GFDL01012808 JAV22237.1 KQ971319 KYB28833.1 ADTU01001874 ADTU01001875 ADTU01001876 ADTU01001877 GBBI01003535 JAC15177.1 GFDL01012782 JAV22263.1 KQ976582 KYM79824.1 EFA00470.2 GBGD01001613 JAC87276.1 KQ414705 KOC63138.1 GEDC01016346 JAS20952.1 KQ982052 KYQ60705.1 GEDC01026806 GEDC01009209 JAS10492.1 JAS28089.1 GL765979 EFZ15760.1 GECL01001406 JAP04718.1 GDHF01023904 JAI28410.1 GAKP01015975 GAKP01015973 GAKP01015972 GAKP01015971 GAKP01015969 JAC42979.1 GDHF01003889 JAI48425.1 GFTR01006031 JAW10395.1 JXJN01004378 JRES01001668 KNC21059.1 GAKP01015976 GAKP01015974 GAKP01015970 GAKP01015968 JAC42982.1 GL888070 EGI68156.1 GAMC01019519 JAB87036.1 GGFJ01004574 MBW53715.1 KQ978739 KYN28856.1 CH933809 KRG05776.1 EDW17805.1 GGFL01003111 MBW67289.1 AXCM01000178 CH940647 KRF84147.1 GGFJ01002685 MBW51826.1 EDW69036.1 GGFK01006170 MBW39491.1 GGFJ01002686 MBW51827.1 GGFK01005911 MBW39232.1 KZ149952 PZC76589.1 KQ978292 KYM95493.1 GANO01000323 JAB59548.1 CH916366 EDV97011.1 AAAB01008960 EAA11394.4 APCN01004428
NWSH01001692 PCG70407.1 KQ459463 KPJ00537.1 KQ435891 KOX69445.1 KQ461108 KPJ09070.1 GEZM01003658 JAV96648.1 GEZM01003660 GEZM01003659 JAV96647.1 JR048067 AEY60571.1 KZ288212 PBC32880.1 KK107119 EZA58596.1 KQ773232 OAD52414.1 KQ434846 KZC08245.1 LBMM01002117 KMQ95253.1 GL435638 EFN72871.1 NNAY01000002 OXU32287.1 GFDF01006743 JAV07341.1 GFDF01006742 JAV07342.1 GBXI01006597 JAD07695.1 GFDF01006647 JAV07437.1 JXUM01040880 JXUM01040881 JXUM01040882 JXUM01040883 JXUM01040884 JXUM01040885 KQ561263 KXJ79163.1 GBXI01013464 GBXI01009827 JAD00828.1 JAD04465.1 GFDL01012808 JAV22237.1 KQ971319 KYB28833.1 ADTU01001874 ADTU01001875 ADTU01001876 ADTU01001877 GBBI01003535 JAC15177.1 GFDL01012782 JAV22263.1 KQ976582 KYM79824.1 EFA00470.2 GBGD01001613 JAC87276.1 KQ414705 KOC63138.1 GEDC01016346 JAS20952.1 KQ982052 KYQ60705.1 GEDC01026806 GEDC01009209 JAS10492.1 JAS28089.1 GL765979 EFZ15760.1 GECL01001406 JAP04718.1 GDHF01023904 JAI28410.1 GAKP01015975 GAKP01015973 GAKP01015972 GAKP01015971 GAKP01015969 JAC42979.1 GDHF01003889 JAI48425.1 GFTR01006031 JAW10395.1 JXJN01004378 JRES01001668 KNC21059.1 GAKP01015976 GAKP01015974 GAKP01015970 GAKP01015968 JAC42982.1 GL888070 EGI68156.1 GAMC01019519 JAB87036.1 GGFJ01004574 MBW53715.1 KQ978739 KYN28856.1 CH933809 KRG05776.1 EDW17805.1 GGFL01003111 MBW67289.1 AXCM01000178 CH940647 KRF84147.1 GGFJ01002685 MBW51826.1 EDW69036.1 GGFK01006170 MBW39491.1 GGFJ01002686 MBW51827.1 GGFK01005911 MBW39232.1 KZ149952 PZC76589.1 KQ978292 KYM95493.1 GANO01000323 JAB59548.1 CH916366 EDV97011.1 AAAB01008960 EAA11394.4 APCN01004428
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053105
UP000053240
+ More
UP000242457 UP000053097 UP000005203 UP000076502 UP000036403 UP000000311 UP000215335 UP000002358 UP000069940 UP000249989 UP000007266 UP000005205 UP000078540 UP000053825 UP000091820 UP000075809 UP000075882 UP000092460 UP000037069 UP000007755 UP000078492 UP000009192 UP000075883 UP000008792 UP000078542 UP000075900 UP000001070 UP000007062 UP000095301 UP000076408 UP000095300 UP000076407 UP000075840 UP000075902 UP000075920
UP000242457 UP000053097 UP000005203 UP000076502 UP000036403 UP000000311 UP000215335 UP000002358 UP000069940 UP000249989 UP000007266 UP000005205 UP000078540 UP000053825 UP000091820 UP000075809 UP000075882 UP000092460 UP000037069 UP000007755 UP000078492 UP000009192 UP000075883 UP000008792 UP000078542 UP000075900 UP000001070 UP000007062 UP000095301 UP000076408 UP000095300 UP000076407 UP000075840 UP000075902 UP000075920
Pfam
PF00102 Y_phosphatase
Interpro
SUPFAM
SSF52799
SSF52799
Gene 3D
ProteinModelPortal
F2Z7K2
H9IXW3
A0A212F9N2
A0A2A4JEA4
A0A194Q4R7
A0A0M8ZR59
+ More
A0A194QVS8 A0A1Y1NGH0 A0A1Y1NLC4 V9IJC8 A0A2A3EMG7 A0A026WSH5 A0A088A9T3 A0A310S4Y0 A0A154P8J7 A0A0J7KY87 E2A118 A0A232FPK1 K7IW23 A0A1L8DLQ0 A0A1L8DLM4 A0A0A1XAE1 A0A1L8DM46 A0A182G3J1 A0A1S4F057 A0A0A1WR00 A0A1Q3F3X2 A0A139WLR4 A0A158NJ25 A0A023F0Y9 A0A1S4F055 A0A1Q3F3Z9 A0A195B6L2 D6WF82 A0A069DU32 A0A0L7QWY6 A0A1B6D5J2 A0A1A9W4A5 A0A151XK75 A0A1B6CAH5 E9IUJ4 A0A0V0G9N3 A0A0K8UPX5 A0A034VKG1 A0A0K8WC51 A0A182KW25 A0A224XD80 A0A1B0AVR3 A0A0L0BPA4 A0A034VLI5 F4WCH6 W8B1A4 A0A2M4BKY4 A0A195EL52 A0A0Q9XB75 B4KZ08 A0A2M4CPT6 A0A182LRF0 A0A0Q9WI44 A0A2M4BFJ4 B4LE80 A0A2M4AFC3 A0A2M4BFM3 A0A2M4AET3 A0A2W1BNF9 A0A195C5P4 A0A182R2L5 U5EYG1 B4J260 Q7PNX1 A0A1I8MBM0 A0A1I8MBM4 A0A182YNM0 A0A1I8NLY9 A0A1I8NLY3 A0A182XD23 A0A182HMF4 A0A182TTC4 A0A182WAL7
A0A194QVS8 A0A1Y1NGH0 A0A1Y1NLC4 V9IJC8 A0A2A3EMG7 A0A026WSH5 A0A088A9T3 A0A310S4Y0 A0A154P8J7 A0A0J7KY87 E2A118 A0A232FPK1 K7IW23 A0A1L8DLQ0 A0A1L8DLM4 A0A0A1XAE1 A0A1L8DM46 A0A182G3J1 A0A1S4F057 A0A0A1WR00 A0A1Q3F3X2 A0A139WLR4 A0A158NJ25 A0A023F0Y9 A0A1S4F055 A0A1Q3F3Z9 A0A195B6L2 D6WF82 A0A069DU32 A0A0L7QWY6 A0A1B6D5J2 A0A1A9W4A5 A0A151XK75 A0A1B6CAH5 E9IUJ4 A0A0V0G9N3 A0A0K8UPX5 A0A034VKG1 A0A0K8WC51 A0A182KW25 A0A224XD80 A0A1B0AVR3 A0A0L0BPA4 A0A034VLI5 F4WCH6 W8B1A4 A0A2M4BKY4 A0A195EL52 A0A0Q9XB75 B4KZ08 A0A2M4CPT6 A0A182LRF0 A0A0Q9WI44 A0A2M4BFJ4 B4LE80 A0A2M4AFC3 A0A2M4BFM3 A0A2M4AET3 A0A2W1BNF9 A0A195C5P4 A0A182R2L5 U5EYG1 B4J260 Q7PNX1 A0A1I8MBM0 A0A1I8MBM4 A0A182YNM0 A0A1I8NLY9 A0A1I8NLY3 A0A182XD23 A0A182HMF4 A0A182TTC4 A0A182WAL7
PDB
5KA1
E-value=2.8687e-88,
Score=830
Ontologies
GO
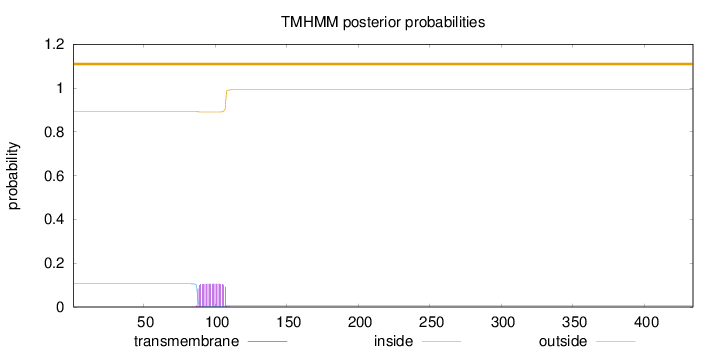
Topology
Length:
434
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.07838
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10570
outside
1 - 434
Population Genetic Test Statistics
Pi
22.308631
Theta
22.369441
Tajima's D
-1.160059
CLR
0.035354
CSRT
0.108594570271486
Interpretation
Uncertain
