Gene
KWMTBOMO00056
Pre Gene Modal
BGIBMGA002032
Annotation
PREDICTED:_mitogen-activated_protein_kinase_kinase_kinase_12-like_[Bombyx_mori]
Full name
Mitogen-activated protein kinase kinase kinase
Location in the cell
Cytoplasmic Reliability : 2.644
Sequence
CDS
ATGGATGAGCACGAGATTGCAGAGAACGAAGCGGGCGGGTTCTTGTCGAAGACTAGTACCCAAGACTTGCGGTTTCACTTCGAAGACGGGAAGAAGCCACTCTTCTGGATGGTGGGAATGATGGACTGCTTCACAAATATTTTCTCGCTCTTCGCCAACAGCGAAATGAGACCAACAAGAGAGGAAGATTGGGAAGTGCCATTTGAGAGCATCACGGATATGATGTACCTAGGCTCTGGGGCCCAGGGGGTAGTGTACAGCGGCAACCTTAGAGGTGAAATGGTAGCCGTCAAAAAATTACGAGAGAAAGAAGAGACCGACATCATGCACCTGAGGAAACTCAACCACGACAACATAGTTCGTTTCCGTGGCGTCTGCTCGGAAGGAGAAAACTATTGTATTATCATGGAGTTCTGTCAATACGGGCCACTCTACGACTTCCTTCACAGCGGTGCATCTTTTACACCCAGACAGATCATTAGGTGGGCCAAAGATATTGCCCAAGGCATGGTCTATTTGCACAACCGCAAGATTATCCATCGTGATCTGAAAAGCCCCAATGTCCTGGTAGCTGACAATCTTGTCGCAAAGGTGAGTGACTTTGGCACCAGCCGAGAGTGGGACAACGAGAGTGCCGTCATGAGCTTTACTGGCACAGTGGCCTGGATGGCGCCCGAAGTGATTCGCCATGAGCCTTGCTCTGAGCGTGTGGATGTCTGGTCTTTCGGTGTTGTGTTATGGGAGCTGTTGACCCAGGAGATCCCGTACAAGACCTTGGAGACTCATGCTATTCTCTGGGGAGTGGGCACCGAAACTATTTTGCTGCCAGTTCCGAACAGTTGCCCGGAGAGCCTGAAACTCTTGCTGAACCAATGCTGGGACCGTGTGCCTAGGAACAGGCCACCTTTCAAGATAATTTTGGCTCACTTGGACATCGCCGGCGCTGAATTCAGTAGGGTCCTGCCCGACCGTTTTAGCGTGACCCAGGCGACATGGCGTAAAGAGATCGAAAATGAAATGAAGGAGATCTATACGAGGAACAAGGAGAAGTTTGAACAGGCTCAGAAACTGCAGTCAGTTCGTCGCGAGGAGTTGCGCTTGGCTCGCGACACGCGTCTCATCTACGAGCAGCAGCTCTCCCGCGCCAACGAACTATACATGGAGGTCTGCGCCGTCCGCCTCCAGCTCGAACAGAAAGAGAAGCACCTTGACGAGCGCGAGAAAGCTCTGAAACTCTGCCGTTGCGGCCTGCGTAAGACTCTGAAGTACTACCAGCGTCAGAACTCGTCATCGTCGGACGGTAAGTCTCCGCTCGAGGTCATCACGGAGGTGCCTCGCCGTCGCAAGCGCCCGGAGGCCCACGTGATCGTGAGCTACATAGAAGAAGACAGACAGACGGTCACCACCGTCATGGAGGAGTGCGAGTGCATAGACAACTGCAAGAACAACAACTTGGACGTAATACAGACCAGCGTGGTCGAGAGGAACGGTAACATTGAAGAGACCACCTGCGCCTTCCCGTTCGGCAGCAATGACAACTACGCCCCAGGTCAAATCCCAGATGTGGCTCACGTCTAA
Protein
MDEHEIAENEAGGFLSKTSTQDLRFHFEDGKKPLFWMVGMMDCFTNIFSLFANSEMRPTREEDWEVPFESITDMMYLGSGAQGVVYSGNLRGEMVAVKKLREKEETDIMHLRKLNHDNIVRFRGVCSEGENYCIIMEFCQYGPLYDFLHSGASFTPRQIIRWAKDIAQGMVYLHNRKIIHRDLKSPNVLVADNLVAKVSDFGTSREWDNESAVMSFTGTVAWMAPEVIRHEPCSERVDVWSFGVVLWELLTQEIPYKTLETHAILWGVGTETILLPVPNSCPESLKLLLNQCWDRVPRNRPPFKIILAHLDIAGAEFSRVLPDRFSVTQATWRKEIENEMKEIYTRNKEKFEQAQKLQSVRREELRLARDTRLIYEQQLSRANELYMEVCAVRLQLEQKEKHLDEREKALKLCRCGLRKTLKYYQRQNSSSSDGKSPLEVITEVPRRRKRPEAHVIVSYIEEDRQTVTTVMEECECIDNCKNNNLDVIQTSVVERNGNIEETTCAFPFGSNDNYAPGQIPDVAHV
Summary
Catalytic Activity
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the protein kinase superfamily. STE Ser/Thr protein kinase family. MAP kinase kinase kinase subfamily.
Belongs to the protein kinase superfamily. STE Ser/Thr protein kinase family. MAP kinase kinase kinase subfamily.
Uniprot
H9IXP9
A0A212F9M5
A0A194Q4S3
A0A194QUD5
A0A2A4K5N2
A0A2W1BSF9
+ More
A0A2H1X228 A0A1B6E331 A0A1B6D4I0 A0A2M3ZGW5 A0A2M3ZGY8 B4H9B4 A0A1B6G767 A0A1B6MNK2 A0A1B6M9P6 A0A1B6M2V8 A0A2H8TQA8 A0A1B6M4E0 A0A2S2QI36 A0A2M4CIW3 A0A2M4A534 A0A2M4A599 A0A2M4CR32 A0A2M4CR65 A0A2M4BAZ4 J9K3N1 A0A0R3P786 Q29DE8 A0A0R3P9A3 T1IM54 A0A3B0J4R0 A0A3B0JTE1 A0A182UAC5 Q16RU6 A0A1W4URS3 A0A1W4UE64 A0A182FF86 A0A0L7QZB4 A0A0Q9WIT7 A0A154PAN9 A0A2A3EKD4 A0A1I8QEA1 B4LD14 A0A087ZUA1 A0A1I8QEC4 A0A0P9ALW0 A0A0P8XVS8 B3M6Y7 A0A0Q9WSY3 A0A2C9JZG5 B4N3L7 A0A0P8XVQ1 A0A224XJB4 A0A210Q125 A0A0M8ZUP8 A0A0R1E0S5 B4IY32 A0A0J9RY47 A0A0Q5U5V1 B4QQW6 B4PGD9 A0A069DWV2 B4IIN8 D3DMF6 B3NIG8 A0A0M4EMX5 Q9VW24 A0A3Q0IUJ5 A0A0Q9XQQ6 E2B5X6 A0A0P4VZ37 B4KZB1 Q7PP29 A0A182MUH1 A0A084VAY7 A0A182X909 K7J705 E9ITI2 A0A1Q3G1Y9 A0A1Q3G1P1 A0A232FB58 A0A182PB91 A0A182LKW7 A0A1I8NHG0 A0A1I8NHE5 A0A0A9YND6 A0A0A9XW76 A0A2J7PN35 C3YYZ9 A0A182QZ51 A0A2T7PX45 A0A2R5LBF1 K1QIA3 A0A2J7PN34 A0A1J1IXK6 A0A2J7PN36 A0A067RB91 A0A195D8B1 V5GTG9 A0A0L0C918
A0A2H1X228 A0A1B6E331 A0A1B6D4I0 A0A2M3ZGW5 A0A2M3ZGY8 B4H9B4 A0A1B6G767 A0A1B6MNK2 A0A1B6M9P6 A0A1B6M2V8 A0A2H8TQA8 A0A1B6M4E0 A0A2S2QI36 A0A2M4CIW3 A0A2M4A534 A0A2M4A599 A0A2M4CR32 A0A2M4CR65 A0A2M4BAZ4 J9K3N1 A0A0R3P786 Q29DE8 A0A0R3P9A3 T1IM54 A0A3B0J4R0 A0A3B0JTE1 A0A182UAC5 Q16RU6 A0A1W4URS3 A0A1W4UE64 A0A182FF86 A0A0L7QZB4 A0A0Q9WIT7 A0A154PAN9 A0A2A3EKD4 A0A1I8QEA1 B4LD14 A0A087ZUA1 A0A1I8QEC4 A0A0P9ALW0 A0A0P8XVS8 B3M6Y7 A0A0Q9WSY3 A0A2C9JZG5 B4N3L7 A0A0P8XVQ1 A0A224XJB4 A0A210Q125 A0A0M8ZUP8 A0A0R1E0S5 B4IY32 A0A0J9RY47 A0A0Q5U5V1 B4QQW6 B4PGD9 A0A069DWV2 B4IIN8 D3DMF6 B3NIG8 A0A0M4EMX5 Q9VW24 A0A3Q0IUJ5 A0A0Q9XQQ6 E2B5X6 A0A0P4VZ37 B4KZB1 Q7PP29 A0A182MUH1 A0A084VAY7 A0A182X909 K7J705 E9ITI2 A0A1Q3G1Y9 A0A1Q3G1P1 A0A232FB58 A0A182PB91 A0A182LKW7 A0A1I8NHG0 A0A1I8NHE5 A0A0A9YND6 A0A0A9XW76 A0A2J7PN35 C3YYZ9 A0A182QZ51 A0A2T7PX45 A0A2R5LBF1 K1QIA3 A0A2J7PN34 A0A1J1IXK6 A0A2J7PN36 A0A067RB91 A0A195D8B1 V5GTG9 A0A0L0C918
EC Number
2.7.11.25
Pubmed
19121390
22118469
26354079
28756777
17994087
15632085
+ More
17510324 18057021 15562597 28812685 17550304 22936249 26334808 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 27129103 12364791 24438588 20075255 21282665 28648823 20966253 25315136 25401762 26823975 18563158 22992520 24845553 25765539 26108605
17510324 18057021 15562597 28812685 17550304 22936249 26334808 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 27129103 12364791 24438588 20075255 21282665 28648823 20966253 25315136 25401762 26823975 18563158 22992520 24845553 25765539 26108605
EMBL
BABH01015735
AGBW02009564
OWR50434.1
KQ459463
KPJ00542.1
KQ461108
+ More
KPJ09077.1 NWSH01000103 PCG79537.1 KZ149952 PZC76584.1 ODYU01012411 SOQ58704.1 GEDC01004983 JAS32315.1 GEDC01016696 JAS20602.1 GGFM01007033 MBW27784.1 GGFM01007010 MBW27761.1 CH479227 EDW35338.1 GECZ01011520 JAS58249.1 GEBQ01002542 JAT37435.1 GEBQ01007336 JAT32641.1 GEBQ01009712 JAT30265.1 GFXV01004384 MBW16189.1 GEBQ01009204 JAT30773.1 GGMS01008174 MBY77377.1 GGFL01001011 MBW65189.1 GGFK01002558 MBW35879.1 GGFK01002666 MBW35987.1 GGFL01003614 MBW67792.1 GGFL01003615 MBW67793.1 GGFJ01001068 MBW50209.1 ABLF02033339 CH379070 KRT08977.1 EAL30466.2 KRT08976.1 JH430968 OUUW01000002 SPP76617.1 SPP76616.1 CH477696 EAT37138.1 KQ414683 KOC63896.1 CH940647 KRF84637.1 KQ434846 KZC08444.1 KZ288223 PBC31954.1 EDW69895.1 CH902618 KPU78814.1 KPU78815.1 EDV40852.1 CH964095 KRF99033.1 EDW79222.1 KPU78816.1 GFTR01007744 JAW08682.1 NEDP02005293 OWF42369.1 KQ435835 KOX71515.1 CM000159 KRK02366.1 CH916366 EDV97575.1 CM002912 KMZ00584.1 CH954178 KQS44254.1 CM000363 EDX11104.1 KMZ00582.1 EDW95298.2 GBGD01000702 JAC88187.1 CH480844 EDW49809.1 BT100072 AE014296 ACX61613.1 ADV37568.1 EDV52393.1 CP012525 ALC45135.1 AY094787 AAF49129.3 AAM11140.1 CH933809 KRG06424.1 GL445887 EFN88945.1 GDKW01001789 JAI54806.1 EDW18937.2 AAAB01008960 EAA11125.5 AXCM01017424 ATLV01004756 ATLV01004757 ATLV01004758 KE524238 KFB35131.1 GL765555 EFZ16110.1 GFDL01001254 JAV33791.1 GFDL01001318 JAV33727.1 NNAY01000580 OXU27567.1 GBHO01011012 GBHO01011011 GDHC01019943 GDHC01018164 GDHC01000698 JAG32592.1 JAG32593.1 JAP98685.1 JAQ00465.1 JAQ17931.1 GBHO01019963 GBHO01011009 GDHC01000094 JAG23641.1 JAG32595.1 JAQ18535.1 NEVH01023957 PNF17751.1 GG666565 EEN54507.1 AXCN02000458 AXCN02000459 AXCN02000460 PZQS01000001 PVD37995.1 GGLE01002696 MBY06822.1 JH817910 EKC28565.1 PNF17752.1 CVRI01000063 CRL04450.1 PNF17750.1 KK852811 KDR15928.1 KQ976750 KYN08679.1 GANP01010758 JAB73710.1 JRES01000836 KNC27874.1
KPJ09077.1 NWSH01000103 PCG79537.1 KZ149952 PZC76584.1 ODYU01012411 SOQ58704.1 GEDC01004983 JAS32315.1 GEDC01016696 JAS20602.1 GGFM01007033 MBW27784.1 GGFM01007010 MBW27761.1 CH479227 EDW35338.1 GECZ01011520 JAS58249.1 GEBQ01002542 JAT37435.1 GEBQ01007336 JAT32641.1 GEBQ01009712 JAT30265.1 GFXV01004384 MBW16189.1 GEBQ01009204 JAT30773.1 GGMS01008174 MBY77377.1 GGFL01001011 MBW65189.1 GGFK01002558 MBW35879.1 GGFK01002666 MBW35987.1 GGFL01003614 MBW67792.1 GGFL01003615 MBW67793.1 GGFJ01001068 MBW50209.1 ABLF02033339 CH379070 KRT08977.1 EAL30466.2 KRT08976.1 JH430968 OUUW01000002 SPP76617.1 SPP76616.1 CH477696 EAT37138.1 KQ414683 KOC63896.1 CH940647 KRF84637.1 KQ434846 KZC08444.1 KZ288223 PBC31954.1 EDW69895.1 CH902618 KPU78814.1 KPU78815.1 EDV40852.1 CH964095 KRF99033.1 EDW79222.1 KPU78816.1 GFTR01007744 JAW08682.1 NEDP02005293 OWF42369.1 KQ435835 KOX71515.1 CM000159 KRK02366.1 CH916366 EDV97575.1 CM002912 KMZ00584.1 CH954178 KQS44254.1 CM000363 EDX11104.1 KMZ00582.1 EDW95298.2 GBGD01000702 JAC88187.1 CH480844 EDW49809.1 BT100072 AE014296 ACX61613.1 ADV37568.1 EDV52393.1 CP012525 ALC45135.1 AY094787 AAF49129.3 AAM11140.1 CH933809 KRG06424.1 GL445887 EFN88945.1 GDKW01001789 JAI54806.1 EDW18937.2 AAAB01008960 EAA11125.5 AXCM01017424 ATLV01004756 ATLV01004757 ATLV01004758 KE524238 KFB35131.1 GL765555 EFZ16110.1 GFDL01001254 JAV33791.1 GFDL01001318 JAV33727.1 NNAY01000580 OXU27567.1 GBHO01011012 GBHO01011011 GDHC01019943 GDHC01018164 GDHC01000698 JAG32592.1 JAG32593.1 JAP98685.1 JAQ00465.1 JAQ17931.1 GBHO01019963 GBHO01011009 GDHC01000094 JAG23641.1 JAG32595.1 JAQ18535.1 NEVH01023957 PNF17751.1 GG666565 EEN54507.1 AXCN02000458 AXCN02000459 AXCN02000460 PZQS01000001 PVD37995.1 GGLE01002696 MBY06822.1 JH817910 EKC28565.1 PNF17752.1 CVRI01000063 CRL04450.1 PNF17750.1 KK852811 KDR15928.1 KQ976750 KYN08679.1 GANP01010758 JAB73710.1 JRES01000836 KNC27874.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000218220
UP000008744
+ More
UP000007819 UP000001819 UP000268350 UP000075902 UP000008820 UP000192221 UP000069272 UP000053825 UP000008792 UP000076502 UP000242457 UP000095300 UP000005203 UP000007801 UP000007798 UP000076420 UP000242188 UP000053105 UP000002282 UP000001070 UP000008711 UP000000304 UP000001292 UP000000803 UP000092553 UP000079169 UP000009192 UP000008237 UP000007062 UP000075883 UP000030765 UP000076407 UP000002358 UP000215335 UP000075885 UP000075882 UP000095301 UP000235965 UP000001554 UP000075886 UP000245119 UP000005408 UP000183832 UP000027135 UP000078542 UP000037069
UP000007819 UP000001819 UP000268350 UP000075902 UP000008820 UP000192221 UP000069272 UP000053825 UP000008792 UP000076502 UP000242457 UP000095300 UP000005203 UP000007801 UP000007798 UP000076420 UP000242188 UP000053105 UP000002282 UP000001070 UP000008711 UP000000304 UP000001292 UP000000803 UP000092553 UP000079169 UP000009192 UP000008237 UP000007062 UP000075883 UP000030765 UP000076407 UP000002358 UP000215335 UP000075885 UP000075882 UP000095301 UP000235965 UP000001554 UP000075886 UP000245119 UP000005408 UP000183832 UP000027135 UP000078542 UP000037069
PRIDE
Interpro
IPR000719
Prot_kinase_dom
+ More
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR008271 Ser/Thr_kinase_AS
IPR013098 Ig_I-set
IPR036116 FN3_sf
IPR003961 FN3_dom
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR017419 MAP3K12_MAP3K13
IPR037278 ARFGAP/RecO
IPR038508 ArfGAP_dom_sf
IPR001164 ArfGAP_dom
IPR017896 4Fe4S_Fe-S-bd
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR008271 Ser/Thr_kinase_AS
IPR013098 Ig_I-set
IPR036116 FN3_sf
IPR003961 FN3_dom
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR017419 MAP3K12_MAP3K13
IPR037278 ARFGAP/RecO
IPR038508 ArfGAP_dom_sf
IPR001164 ArfGAP_dom
IPR017896 4Fe4S_Fe-S-bd
Gene 3D
CDD
ProteinModelPortal
H9IXP9
A0A212F9M5
A0A194Q4S3
A0A194QUD5
A0A2A4K5N2
A0A2W1BSF9
+ More
A0A2H1X228 A0A1B6E331 A0A1B6D4I0 A0A2M3ZGW5 A0A2M3ZGY8 B4H9B4 A0A1B6G767 A0A1B6MNK2 A0A1B6M9P6 A0A1B6M2V8 A0A2H8TQA8 A0A1B6M4E0 A0A2S2QI36 A0A2M4CIW3 A0A2M4A534 A0A2M4A599 A0A2M4CR32 A0A2M4CR65 A0A2M4BAZ4 J9K3N1 A0A0R3P786 Q29DE8 A0A0R3P9A3 T1IM54 A0A3B0J4R0 A0A3B0JTE1 A0A182UAC5 Q16RU6 A0A1W4URS3 A0A1W4UE64 A0A182FF86 A0A0L7QZB4 A0A0Q9WIT7 A0A154PAN9 A0A2A3EKD4 A0A1I8QEA1 B4LD14 A0A087ZUA1 A0A1I8QEC4 A0A0P9ALW0 A0A0P8XVS8 B3M6Y7 A0A0Q9WSY3 A0A2C9JZG5 B4N3L7 A0A0P8XVQ1 A0A224XJB4 A0A210Q125 A0A0M8ZUP8 A0A0R1E0S5 B4IY32 A0A0J9RY47 A0A0Q5U5V1 B4QQW6 B4PGD9 A0A069DWV2 B4IIN8 D3DMF6 B3NIG8 A0A0M4EMX5 Q9VW24 A0A3Q0IUJ5 A0A0Q9XQQ6 E2B5X6 A0A0P4VZ37 B4KZB1 Q7PP29 A0A182MUH1 A0A084VAY7 A0A182X909 K7J705 E9ITI2 A0A1Q3G1Y9 A0A1Q3G1P1 A0A232FB58 A0A182PB91 A0A182LKW7 A0A1I8NHG0 A0A1I8NHE5 A0A0A9YND6 A0A0A9XW76 A0A2J7PN35 C3YYZ9 A0A182QZ51 A0A2T7PX45 A0A2R5LBF1 K1QIA3 A0A2J7PN34 A0A1J1IXK6 A0A2J7PN36 A0A067RB91 A0A195D8B1 V5GTG9 A0A0L0C918
A0A2H1X228 A0A1B6E331 A0A1B6D4I0 A0A2M3ZGW5 A0A2M3ZGY8 B4H9B4 A0A1B6G767 A0A1B6MNK2 A0A1B6M9P6 A0A1B6M2V8 A0A2H8TQA8 A0A1B6M4E0 A0A2S2QI36 A0A2M4CIW3 A0A2M4A534 A0A2M4A599 A0A2M4CR32 A0A2M4CR65 A0A2M4BAZ4 J9K3N1 A0A0R3P786 Q29DE8 A0A0R3P9A3 T1IM54 A0A3B0J4R0 A0A3B0JTE1 A0A182UAC5 Q16RU6 A0A1W4URS3 A0A1W4UE64 A0A182FF86 A0A0L7QZB4 A0A0Q9WIT7 A0A154PAN9 A0A2A3EKD4 A0A1I8QEA1 B4LD14 A0A087ZUA1 A0A1I8QEC4 A0A0P9ALW0 A0A0P8XVS8 B3M6Y7 A0A0Q9WSY3 A0A2C9JZG5 B4N3L7 A0A0P8XVQ1 A0A224XJB4 A0A210Q125 A0A0M8ZUP8 A0A0R1E0S5 B4IY32 A0A0J9RY47 A0A0Q5U5V1 B4QQW6 B4PGD9 A0A069DWV2 B4IIN8 D3DMF6 B3NIG8 A0A0M4EMX5 Q9VW24 A0A3Q0IUJ5 A0A0Q9XQQ6 E2B5X6 A0A0P4VZ37 B4KZB1 Q7PP29 A0A182MUH1 A0A084VAY7 A0A182X909 K7J705 E9ITI2 A0A1Q3G1Y9 A0A1Q3G1P1 A0A232FB58 A0A182PB91 A0A182LKW7 A0A1I8NHG0 A0A1I8NHE5 A0A0A9YND6 A0A0A9XW76 A0A2J7PN35 C3YYZ9 A0A182QZ51 A0A2T7PX45 A0A2R5LBF1 K1QIA3 A0A2J7PN34 A0A1J1IXK6 A0A2J7PN36 A0A067RB91 A0A195D8B1 V5GTG9 A0A0L0C918
Ontologies
GO
Topology
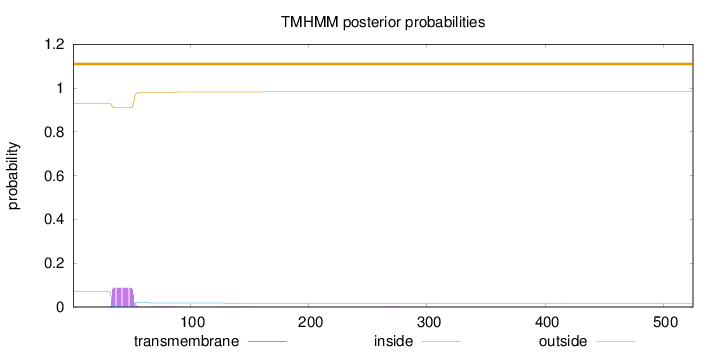
Length:
525
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.759
Exp number, first 60 AAs:
1.68751
Total prob of N-in:
0.07018
outside
1 - 525
Population Genetic Test Statistics
Pi
21.623573
Theta
27.431769
Tajima's D
-0.529058
CLR
0.021236
CSRT
0.235838208089596
Interpretation
Uncertain
