Gene
KWMTBOMO00052
Pre Gene Modal
BGIBMGA002093
Annotation
PREDICTED:_solute_carrier_family_35_member_C2_[Bombyx_mori]
Full name
Ubiquitin carboxyl-terminal hydrolase
Location in the cell
PlasmaMembrane Reliability : 4.934
Sequence
CDS
ATGTACATACAAATTTTCACAAAAATAAGAATTGTTATTGTTTTATTTTCTCATTGTACTGAGAAGAAGGACTTCAAATACCCATTGACGGTGGTGATGTACCACTTGGTTGTGAAGTGGTTGCTGTCTGTGCTAGTGCGTTCCGTGCTGTACTGCATCACGAAGACCCCTCAGCTGGTGTTGCCTCTCGGGACTAGCATCCGGTCCGTGGCTCCGACCGGCCTGTCCAGCGGCATCGACGTGGGCTTCTCGAATTGGGGCCTGGAGCTCGTCACCATATCGCTGTACACAATGACGAAATCGACAACGATCATTTTCATTCTCGGCTTCGCGATAATTCTCGGTTTGGAAAAGAAGTCTTGGTCTCTCGTGGGCATCGTCCTCATGATCGCGGCGGGCCTTATCATGTTTACTTACAAAGCGACCCAGTTCAACATCGAAGGATTCAGTTTTCTACTCCTAGCGTCTTTTGCGGCGGGTCTACGTTGGACCTTCGCTCAACTTCTCATGCAGAAGGCGAAGCTGGGACTCCATAACCCAGTGGATATGGTGTTTCACGTGCAGCCTTGGATGTTTTTAGCCCTGATACCATTTACAGCAATTTTTGAAGGCCTGGATTGCATCATGTTCCTACTAGGTCTACCCGACGAGTTGTTACTCCCAACTGTCTTGAAGGTGACCGTTGGAGCCACGATCGCATTCGCAATGGAGATCAGCGAGTTCCTGGTCGTAACCTACACTTCGAGTCTGACGCTGTCCATTGCTGGAATATTCAAGGAGATGTGCATACTGGTCTTAGCGGTGAAGGTGAGCGGCGATCAGCTCAGCCTCATAAACGTGGTCGGTCTTGCGGTGTGTCTGCTCGGGATCATTGGCCACATCGTCCATAAGATGTTGATCATTAGAGCCGTCGAGAGTGCCGTTGTTGTCGAAGTCGATAACTTCGAGGCTGTCAAGAACAGGTCCCCGAATAAAGACGAAACAGATGAACCTCTACTGACGGAGAAGAGTTGGCTTAGCGAAGACAGCGACGTAGACGCTAACGTGGTGCTATACGAAGTATTGCAGAGACGCGACGGACATTAA
Protein
MYIQIFTKIRIVIVLFSHCTEKKDFKYPLTVVMYHLVVKWLLSVLVRSVLYCITKTPQLVLPLGTSIRSVAPTGLSSGIDVGFSNWGLELVTISLYTMTKSTTIIFILGFAIILGLEKKSWSLVGIVLMIAAGLIMFTYKATQFNIEGFSFLLLASFAAGLRWTFAQLLMQKAKLGLHNPVDMVFHVQPWMFLALIPFTAIFEGLDCIMFLLGLPDELLLPTVLKVTVGATIAFAMEISEFLVVTYTSSLTLSIAGIFKEMCILVLAVKVSGDQLSLINVVGLAVCLLGIIGHIVHKMLIIRAVESAVVVEVDNFEAVKNRSPNKDETDEPLLTEKSWLSEDSDVDANVVLYEVLQRRDGH
Summary
Catalytic Activity
Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
Similarity
Belongs to the peptidase C12 family.
Uniprot
A0A2A4IUB8
A0A2A4IUY0
A0A2W1BSH0
A0A1E1W192
A0A212F9M1
A0A194QU69
+ More
A0A194Q6A7 H9IXW0 A0A084WKJ4 A0A182HA21 A0A023ESW0 Q16HA4 Q16J20 A0A0K8TQP9 A0A182PP50 A0A182ITB7 A0A182NM34 A0A182Y106 A0A182TYT1 A0A182WQW6 A0A182JY26 A0A182VDG2 A0A182M0I5 A0A2C9GQQ1 Q7PNQ1 U5EVP5 A0A182QZ50 B0WNW5 A0A182X544 A0A1Q3FI62 A0A1Q3FIA8 A0A1Q3FI66 A0A2J7Q3H9 A0A2M4BK32 A0A2M4BKT5 A0A182RN10 A0A067QV19 A0A2M4AIG5 A0A2M3Z6L7 W5J9I1 A0A1A9YS24 A0A1B0AE06 A0A336KUE2 A0A336M8J8 A0A1A9WCR2 A0A1L8DDG8 A0A1B0FR02 A0A1L8DDP3 A0A1L8DDP1 A0A1L8E1M3 A0A1L8E151 A0A1I8P7A5 A0A1I8MXL5 A0A1L8DDG5 A0A182FHX4 A0A1L8E190 B4LH38 A0A1W4X8Z7 A0A0L0BM24 B4KWE7 E0VZY3 A0A1B0B8N5 B4N570 B4IXC6 A0A158NAM5 A0A151I5V5 B4PH41 B3NBX3 A0A0V0G906 E2ANR2 E9IPJ4 A0A1W4W0I8 A0A023F9R1 A0A026WQY0 A0A1Y1KHE0 B3M827 Q960F0 Q9VZP2 E2B5X8 A0A195CJG5 A0A0P4VHU2 B4QPJ6 R4G304 A0A0H3YFJ8 A0A0A1WYV7 B7PD55 A0A1J1ISQ4 D6WIN5 A0A034WBH3 A0A0K8VJC4 A0A1B0GID3 A0A3B0K1P1 A0A232FAT0 A0A1D2NCU3 A0A154P9C4 Q29DQ0 A0A0L7QZ70 K7J704 A0A1W4XIG8 W8CDA6
A0A194Q6A7 H9IXW0 A0A084WKJ4 A0A182HA21 A0A023ESW0 Q16HA4 Q16J20 A0A0K8TQP9 A0A182PP50 A0A182ITB7 A0A182NM34 A0A182Y106 A0A182TYT1 A0A182WQW6 A0A182JY26 A0A182VDG2 A0A182M0I5 A0A2C9GQQ1 Q7PNQ1 U5EVP5 A0A182QZ50 B0WNW5 A0A182X544 A0A1Q3FI62 A0A1Q3FIA8 A0A1Q3FI66 A0A2J7Q3H9 A0A2M4BK32 A0A2M4BKT5 A0A182RN10 A0A067QV19 A0A2M4AIG5 A0A2M3Z6L7 W5J9I1 A0A1A9YS24 A0A1B0AE06 A0A336KUE2 A0A336M8J8 A0A1A9WCR2 A0A1L8DDG8 A0A1B0FR02 A0A1L8DDP3 A0A1L8DDP1 A0A1L8E1M3 A0A1L8E151 A0A1I8P7A5 A0A1I8MXL5 A0A1L8DDG5 A0A182FHX4 A0A1L8E190 B4LH38 A0A1W4X8Z7 A0A0L0BM24 B4KWE7 E0VZY3 A0A1B0B8N5 B4N570 B4IXC6 A0A158NAM5 A0A151I5V5 B4PH41 B3NBX3 A0A0V0G906 E2ANR2 E9IPJ4 A0A1W4W0I8 A0A023F9R1 A0A026WQY0 A0A1Y1KHE0 B3M827 Q960F0 Q9VZP2 E2B5X8 A0A195CJG5 A0A0P4VHU2 B4QPJ6 R4G304 A0A0H3YFJ8 A0A0A1WYV7 B7PD55 A0A1J1ISQ4 D6WIN5 A0A034WBH3 A0A0K8VJC4 A0A1B0GID3 A0A3B0K1P1 A0A232FAT0 A0A1D2NCU3 A0A154P9C4 Q29DQ0 A0A0L7QZ70 K7J704 A0A1W4XIG8 W8CDA6
EC Number
3.4.19.12
Pubmed
28756777
22118469
26354079
19121390
24438588
26483478
+ More
24945155 17510324 26369729 25244985 12364791 14747013 17210077 24845553 20920257 23761445 25315136 17994087 26108605 18057021 20566863 21347285 17550304 20798317 21282665 25474469 24508170 30249741 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 22936249 26057828 25830018 18362917 19820115 25348373 28648823 27289101 15632085 23185243 20075255 24495485
24945155 17510324 26369729 25244985 12364791 14747013 17210077 24845553 20920257 23761445 25315136 17994087 26108605 18057021 20566863 21347285 17550304 20798317 21282665 25474469 24508170 30249741 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 22936249 26057828 25830018 18362917 19820115 25348373 28648823 27289101 15632085 23185243 20075255 24495485
EMBL
NWSH01006238
PCG63577.1
PCG63575.1
KZ149952
PZC76575.1
GDQN01010352
+ More
JAT80702.1 AGBW02009564 OWR50430.1 KQ461108 KPJ09073.1 KQ459463 KPJ00540.1 BABH01015712 ATLV01024123 KE525349 KFB50738.1 JXUM01121934 KQ566552 KXJ70039.1 GAPW01001221 JAC12377.1 CH478188 EAT33620.1 CH478036 EAT34267.1 GDAI01000916 JAI16687.1 AXCM01013109 APCN01004280 APCN01004281 AAAB01008960 EAA11804.4 GANO01003316 JAB56555.1 AXCN02002327 DS232016 EDS31970.1 GFDL01007798 JAV27247.1 GFDL01007767 JAV27278.1 GFDL01007810 JAV27235.1 NEVH01019067 PNF23148.1 GGFJ01004266 MBW53407.1 GGFJ01004267 MBW53408.1 KK852982 KDR12889.1 GGFK01007216 MBW40537.1 GGFM01003422 MBW24173.1 ADMH02001883 ETN60656.1 UFQS01000972 UFQT01000972 SSX08095.1 SSX28235.1 UFQT01000553 SSX25219.1 GFDF01009588 JAV04496.1 CCAG010001599 GFDF01009587 JAV04497.1 GFDF01009589 JAV04495.1 GFDF01001612 JAV12472.1 GFDF01001611 JAV12473.1 GFDF01009583 JAV04501.1 GFDF01001613 JAV12471.1 CH940647 EDW69528.1 JRES01001656 KNC21101.1 CH933809 EDW18554.1 KRG06245.1 DS235854 EEB18939.1 JXJN01010019 CH964101 EDW79509.1 CH916366 EDV97458.1 ADTU01010381 KQ976413 KYM90357.1 CM000159 EDW93278.1 CH954178 EDV50790.1 GECL01001606 JAP04518.1 GL441305 EFN64930.1 GL764522 EFZ17518.1 GBBI01001049 JAC17663.1 KK107145 QOIP01000010 EZA57504.1 RLU17423.1 RLU17644.1 GEZM01083974 JAV60889.1 CH902618 EDV39935.1 AY052095 AAK93519.1 AE014296 BT133405 AAF47777.1 AFH55504.1 GL445887 EFN88947.1 KQ977642 KYN00881.1 GDKW01002358 JAI54237.1 CM000363 CM002912 EDX09089.1 KMY97369.1 ACPB03001509 GAHY01002121 JAA75389.1 KT163697 AKN21647.1 GBXI01010048 JAD04244.1 ABJB010047318 ABJB010332044 ABJB010641635 ABJB010825748 ABJB010886116 DS688408 EEC04527.1 CVRI01000059 CRL03160.1 KQ971321 EFA00757.1 GAKP01006026 JAC52926.1 GDHF01013325 JAI38989.1 AJWK01013699 AJWK01013700 AJWK01013701 AJWK01013702 AJWK01013703 AJWK01013704 AJWK01013705 AJWK01013706 OUUW01000012 SPP87233.1 NNAY01000580 OXU27569.1 LJIJ01000095 ODN02816.1 KQ434846 KZC08441.1 CH379070 EAL30364.1 KRT08861.1 KQ414683 KOC63893.1 GAMC01002281 JAC04275.1
JAT80702.1 AGBW02009564 OWR50430.1 KQ461108 KPJ09073.1 KQ459463 KPJ00540.1 BABH01015712 ATLV01024123 KE525349 KFB50738.1 JXUM01121934 KQ566552 KXJ70039.1 GAPW01001221 JAC12377.1 CH478188 EAT33620.1 CH478036 EAT34267.1 GDAI01000916 JAI16687.1 AXCM01013109 APCN01004280 APCN01004281 AAAB01008960 EAA11804.4 GANO01003316 JAB56555.1 AXCN02002327 DS232016 EDS31970.1 GFDL01007798 JAV27247.1 GFDL01007767 JAV27278.1 GFDL01007810 JAV27235.1 NEVH01019067 PNF23148.1 GGFJ01004266 MBW53407.1 GGFJ01004267 MBW53408.1 KK852982 KDR12889.1 GGFK01007216 MBW40537.1 GGFM01003422 MBW24173.1 ADMH02001883 ETN60656.1 UFQS01000972 UFQT01000972 SSX08095.1 SSX28235.1 UFQT01000553 SSX25219.1 GFDF01009588 JAV04496.1 CCAG010001599 GFDF01009587 JAV04497.1 GFDF01009589 JAV04495.1 GFDF01001612 JAV12472.1 GFDF01001611 JAV12473.1 GFDF01009583 JAV04501.1 GFDF01001613 JAV12471.1 CH940647 EDW69528.1 JRES01001656 KNC21101.1 CH933809 EDW18554.1 KRG06245.1 DS235854 EEB18939.1 JXJN01010019 CH964101 EDW79509.1 CH916366 EDV97458.1 ADTU01010381 KQ976413 KYM90357.1 CM000159 EDW93278.1 CH954178 EDV50790.1 GECL01001606 JAP04518.1 GL441305 EFN64930.1 GL764522 EFZ17518.1 GBBI01001049 JAC17663.1 KK107145 QOIP01000010 EZA57504.1 RLU17423.1 RLU17644.1 GEZM01083974 JAV60889.1 CH902618 EDV39935.1 AY052095 AAK93519.1 AE014296 BT133405 AAF47777.1 AFH55504.1 GL445887 EFN88947.1 KQ977642 KYN00881.1 GDKW01002358 JAI54237.1 CM000363 CM002912 EDX09089.1 KMY97369.1 ACPB03001509 GAHY01002121 JAA75389.1 KT163697 AKN21647.1 GBXI01010048 JAD04244.1 ABJB010047318 ABJB010332044 ABJB010641635 ABJB010825748 ABJB010886116 DS688408 EEC04527.1 CVRI01000059 CRL03160.1 KQ971321 EFA00757.1 GAKP01006026 JAC52926.1 GDHF01013325 JAI38989.1 AJWK01013699 AJWK01013700 AJWK01013701 AJWK01013702 AJWK01013703 AJWK01013704 AJWK01013705 AJWK01013706 OUUW01000012 SPP87233.1 NNAY01000580 OXU27569.1 LJIJ01000095 ODN02816.1 KQ434846 KZC08441.1 CH379070 EAL30364.1 KRT08861.1 KQ414683 KOC63893.1 GAMC01002281 JAC04275.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000005204
UP000030765
+ More
UP000069940 UP000249989 UP000008820 UP000075885 UP000075880 UP000075884 UP000076408 UP000075902 UP000075920 UP000075881 UP000075903 UP000075883 UP000075840 UP000007062 UP000075886 UP000002320 UP000076407 UP000235965 UP000075900 UP000027135 UP000000673 UP000092443 UP000092445 UP000091820 UP000092444 UP000095300 UP000095301 UP000069272 UP000008792 UP000192223 UP000037069 UP000009192 UP000009046 UP000092460 UP000007798 UP000001070 UP000005205 UP000078540 UP000002282 UP000008711 UP000000311 UP000192221 UP000053097 UP000279307 UP000007801 UP000000803 UP000008237 UP000078542 UP000000304 UP000015103 UP000001555 UP000183832 UP000007266 UP000092461 UP000268350 UP000215335 UP000094527 UP000076502 UP000001819 UP000053825 UP000002358
UP000069940 UP000249989 UP000008820 UP000075885 UP000075880 UP000075884 UP000076408 UP000075902 UP000075920 UP000075881 UP000075903 UP000075883 UP000075840 UP000007062 UP000075886 UP000002320 UP000076407 UP000235965 UP000075900 UP000027135 UP000000673 UP000092443 UP000092445 UP000091820 UP000092444 UP000095300 UP000095301 UP000069272 UP000008792 UP000192223 UP000037069 UP000009192 UP000009046 UP000092460 UP000007798 UP000001070 UP000005205 UP000078540 UP000002282 UP000008711 UP000000311 UP000192221 UP000053097 UP000279307 UP000007801 UP000000803 UP000008237 UP000078542 UP000000304 UP000015103 UP000001555 UP000183832 UP000007266 UP000092461 UP000268350 UP000215335 UP000094527 UP000076502 UP000001819 UP000053825 UP000002358
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2A4IUB8
A0A2A4IUY0
A0A2W1BSH0
A0A1E1W192
A0A212F9M1
A0A194QU69
+ More
A0A194Q6A7 H9IXW0 A0A084WKJ4 A0A182HA21 A0A023ESW0 Q16HA4 Q16J20 A0A0K8TQP9 A0A182PP50 A0A182ITB7 A0A182NM34 A0A182Y106 A0A182TYT1 A0A182WQW6 A0A182JY26 A0A182VDG2 A0A182M0I5 A0A2C9GQQ1 Q7PNQ1 U5EVP5 A0A182QZ50 B0WNW5 A0A182X544 A0A1Q3FI62 A0A1Q3FIA8 A0A1Q3FI66 A0A2J7Q3H9 A0A2M4BK32 A0A2M4BKT5 A0A182RN10 A0A067QV19 A0A2M4AIG5 A0A2M3Z6L7 W5J9I1 A0A1A9YS24 A0A1B0AE06 A0A336KUE2 A0A336M8J8 A0A1A9WCR2 A0A1L8DDG8 A0A1B0FR02 A0A1L8DDP3 A0A1L8DDP1 A0A1L8E1M3 A0A1L8E151 A0A1I8P7A5 A0A1I8MXL5 A0A1L8DDG5 A0A182FHX4 A0A1L8E190 B4LH38 A0A1W4X8Z7 A0A0L0BM24 B4KWE7 E0VZY3 A0A1B0B8N5 B4N570 B4IXC6 A0A158NAM5 A0A151I5V5 B4PH41 B3NBX3 A0A0V0G906 E2ANR2 E9IPJ4 A0A1W4W0I8 A0A023F9R1 A0A026WQY0 A0A1Y1KHE0 B3M827 Q960F0 Q9VZP2 E2B5X8 A0A195CJG5 A0A0P4VHU2 B4QPJ6 R4G304 A0A0H3YFJ8 A0A0A1WYV7 B7PD55 A0A1J1ISQ4 D6WIN5 A0A034WBH3 A0A0K8VJC4 A0A1B0GID3 A0A3B0K1P1 A0A232FAT0 A0A1D2NCU3 A0A154P9C4 Q29DQ0 A0A0L7QZ70 K7J704 A0A1W4XIG8 W8CDA6
A0A194Q6A7 H9IXW0 A0A084WKJ4 A0A182HA21 A0A023ESW0 Q16HA4 Q16J20 A0A0K8TQP9 A0A182PP50 A0A182ITB7 A0A182NM34 A0A182Y106 A0A182TYT1 A0A182WQW6 A0A182JY26 A0A182VDG2 A0A182M0I5 A0A2C9GQQ1 Q7PNQ1 U5EVP5 A0A182QZ50 B0WNW5 A0A182X544 A0A1Q3FI62 A0A1Q3FIA8 A0A1Q3FI66 A0A2J7Q3H9 A0A2M4BK32 A0A2M4BKT5 A0A182RN10 A0A067QV19 A0A2M4AIG5 A0A2M3Z6L7 W5J9I1 A0A1A9YS24 A0A1B0AE06 A0A336KUE2 A0A336M8J8 A0A1A9WCR2 A0A1L8DDG8 A0A1B0FR02 A0A1L8DDP3 A0A1L8DDP1 A0A1L8E1M3 A0A1L8E151 A0A1I8P7A5 A0A1I8MXL5 A0A1L8DDG5 A0A182FHX4 A0A1L8E190 B4LH38 A0A1W4X8Z7 A0A0L0BM24 B4KWE7 E0VZY3 A0A1B0B8N5 B4N570 B4IXC6 A0A158NAM5 A0A151I5V5 B4PH41 B3NBX3 A0A0V0G906 E2ANR2 E9IPJ4 A0A1W4W0I8 A0A023F9R1 A0A026WQY0 A0A1Y1KHE0 B3M827 Q960F0 Q9VZP2 E2B5X8 A0A195CJG5 A0A0P4VHU2 B4QPJ6 R4G304 A0A0H3YFJ8 A0A0A1WYV7 B7PD55 A0A1J1ISQ4 D6WIN5 A0A034WBH3 A0A0K8VJC4 A0A1B0GID3 A0A3B0K1P1 A0A232FAT0 A0A1D2NCU3 A0A154P9C4 Q29DQ0 A0A0L7QZ70 K7J704 A0A1W4XIG8 W8CDA6
Ontologies
KEGG
GO
PANTHER
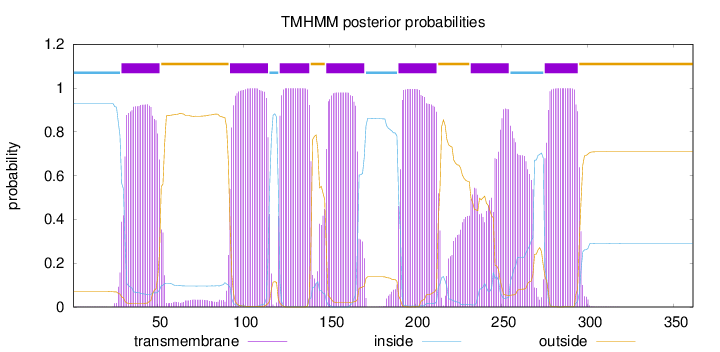
Topology
Length:
361
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
155.57462
Exp number, first 60 AAs:
20.45578
Total prob of N-in:
0.93021
POSSIBLE N-term signal
sequence
inside
1 - 28
TMhelix
29 - 51
outside
52 - 91
TMhelix
92 - 114
inside
115 - 120
TMhelix
121 - 138
outside
139 - 147
TMhelix
148 - 170
inside
171 - 189
TMhelix
190 - 212
outside
213 - 231
TMhelix
232 - 254
inside
255 - 274
TMhelix
275 - 294
outside
295 - 361
Population Genetic Test Statistics
Pi
25.280549
Theta
29.310522
Tajima's D
-0.713909
CLR
0.017631
CSRT
0.190140492975351
Interpretation
Uncertain
