Pre Gene Modal
BGIBMGA002039
Annotation
gamma-soluble_nsf_attachment_protein_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.671 Nuclear Reliability : 1.902
Sequence
CDS
ATGCGTTTGATTGCGAAGGACGCTGCTCGGCCTCTGTTCAACTTGGCACTGGAGGCCTCTAGCCTGTATCAGCAGCACGGGTCCGGGGACAGCGCCGCCGGACTCTTGGACAAGGCCGGTCGCATCCTAGAGCAGGACACACCCCAGCTGGCCGTGAAGCTGTACCAGCACGCTGCCGATATTTCTGCCAACGAGAGCAGCCAGCATCAGGGCATCGAGTACATAAGCAAGGCGTCCAGATTGTTGGTTCGTCTCGAACGCTACGACGAGGCTGTCGATAATTTGCGTCGCGAGATCGGGTTCCATCTTGAAGCGAATAACTTGCCGGCCGTGGGCAGACTGTGCGTGGCCATCGTGCTGGTACAACTGGCCCGCGGCGATACCGTCGCTGCCGAGAAGAGCTACAAGGAATGGGGCGGTCACTGCGAAATGGCCGAAGTGCAGACGTTGGATCAGCTGCTCCAGGCCTACGATGAAGAGGACCCGGATTCCGCTCGCAAGGCCCTCCGTTCCCCGTTCATCCGCAGCATGGACGTGGAGTACTCCCGCCTGGCTAACTGCATCCCGCTTCCGGAGCCACTGGACCCGATGATGCGGGCCGGAGTGCGAGAGAACGCCGCGCCGTCGTACGTCAGCCCTAACGCCAGTACATCCGCCTCGGTGGAACATCATCGTGCCTACGACGAGGAGAATGAGGAATCTCCGTATGAGCCCGTCACTTACGGCCCCAAGAAGATCGAGGAAGAGGACGAAGACGAGGAGCTCTGCTAA
Protein
MRLIAKDAARPLFNLALEASSLYQQHGSGDSAAGLLDKAGRILEQDTPQLAVKLYQHAADISANESSQHQGIEYISKASRLLVRLERYDEAVDNLRREIGFHLEANNLPAVGRLCVAIVLVQLARGDTVAAEKSYKEWGGHCEMAEVQTLDQLLQAYDEEDPDSARKALRSPFIRSMDVEYSRLANCIPLPEPLDPMMRAGVRENAAPSYVSPNASTSASVEHHRAYDEENEESPYEPVTYGPKKIEEEDEDEELC
Summary
Uniprot
H9IXQ6
A0A2W1BBA7
A0A2A4JAT7
A0A212EQM0
A0A194PTV9
A0A194QZM9
+ More
A0A2J7PL56 A0A2J7PL85 A0A067R1P7 A0A2P8YWW5 A0A1Y1N4C4 A0A1B6GL30 E0W1T6 D6WIT9 A0A2M3ZIL5 A0A182Q2P4 W5JD93 A0A182WB39 A0A2M4BU71 A0A2M4AQB4 A0A182RF82 T1DTD4 A0A154PLN7 A0A336MX58 A0A182FDN5 A0A182NGU9 A0A182VP55 E2BSU7 Q7Q6W8 A0A182XMY3 A0A182LJ62 A0A182MUH8 A0A232F1B1 K7J6B2 A0A182PHL0 A0A182Y189 A0A084W9Z0 B0WC92 A0A023EPJ3 A0A182GG38 A0A182J0F2 Q16SG8 A0A0M8ZVC9 U5EYE1 E9J7J4 A0A0J7KX45 A0A182G2J5 A0A1Q3FGW5 A0A182TYS6 A0A1I8NDJ5 A0A2A3E442 V9IKI0 A0A088A5R0 T1PC55 A0A151JLL2 A0A158NYZ3 A0A034WV86 A0A0C9S0H5 A0A151I5C6 F4W554 A0A0K8TSX6 A0A182HWT7 A0A1B6CXK6 A0A026W7R2 E2AN98 A0A195D0U7 A0A1I8PG99 A0A224XSA5 A0A2R7VT43 T1IE43 A0A0A9YF54 A0A146LWN8 A0A0N7Z8N6 A0A1J1J823 A0A069DSW4 A0A1S4EGW3 A0A023F1Z5 A0A0V0G6D3 A0A182JV78 A0A310SBX5 A0A151WQL2 A0A226EWY4 A0A1A9WP66 A0A1B6EA46 A0A195FSM5 A0A240SX67 D3TKN6 A0A240SWW5 A0A1A9Y6J6 A0A1B0BIB0 A0A1A9UJ22 A0A0K8W9U2 A0A0L7R9N8 W8C2Z8 A0A0A1WWM4 W8C7Z4
A0A2J7PL56 A0A2J7PL85 A0A067R1P7 A0A2P8YWW5 A0A1Y1N4C4 A0A1B6GL30 E0W1T6 D6WIT9 A0A2M3ZIL5 A0A182Q2P4 W5JD93 A0A182WB39 A0A2M4BU71 A0A2M4AQB4 A0A182RF82 T1DTD4 A0A154PLN7 A0A336MX58 A0A182FDN5 A0A182NGU9 A0A182VP55 E2BSU7 Q7Q6W8 A0A182XMY3 A0A182LJ62 A0A182MUH8 A0A232F1B1 K7J6B2 A0A182PHL0 A0A182Y189 A0A084W9Z0 B0WC92 A0A023EPJ3 A0A182GG38 A0A182J0F2 Q16SG8 A0A0M8ZVC9 U5EYE1 E9J7J4 A0A0J7KX45 A0A182G2J5 A0A1Q3FGW5 A0A182TYS6 A0A1I8NDJ5 A0A2A3E442 V9IKI0 A0A088A5R0 T1PC55 A0A151JLL2 A0A158NYZ3 A0A034WV86 A0A0C9S0H5 A0A151I5C6 F4W554 A0A0K8TSX6 A0A182HWT7 A0A1B6CXK6 A0A026W7R2 E2AN98 A0A195D0U7 A0A1I8PG99 A0A224XSA5 A0A2R7VT43 T1IE43 A0A0A9YF54 A0A146LWN8 A0A0N7Z8N6 A0A1J1J823 A0A069DSW4 A0A1S4EGW3 A0A023F1Z5 A0A0V0G6D3 A0A182JV78 A0A310SBX5 A0A151WQL2 A0A226EWY4 A0A1A9WP66 A0A1B6EA46 A0A195FSM5 A0A240SX67 D3TKN6 A0A240SWW5 A0A1A9Y6J6 A0A1B0BIB0 A0A1A9UJ22 A0A0K8W9U2 A0A0L7R9N8 W8C2Z8 A0A0A1WWM4 W8C7Z4
Pubmed
19121390
28756777
22118469
26354079
24845553
29403074
+ More
28004739 20566863 18362917 19820115 20920257 23761445 20798317 12364791 14747013 17210077 20966253 28648823 20075255 25244985 24438588 24945155 26483478 17510324 21282665 25315136 21347285 25348373 21719571 26369729 24508170 30249741 25401762 26823975 27129103 26334808 25474469 20353571 24495485 25830018
28004739 20566863 18362917 19820115 20920257 23761445 20798317 12364791 14747013 17210077 20966253 28648823 20075255 25244985 24438588 24945155 26483478 17510324 21282665 25315136 21347285 25348373 21719571 26369729 24508170 30249741 25401762 26823975 27129103 26334808 25474469 20353571 24495485 25830018
EMBL
BABH01015695
BABH01015696
BABH01015697
BABH01015698
BABH01015699
BABH01015700
+ More
BABH01015701 KZ150173 PZC72592.1 NWSH01002101 PCG69195.1 AGBW02013268 OWR43790.1 KQ459593 KPI96752.1 KQ461108 KPJ09051.1 NEVH01024428 PNF17070.1 PNF17069.1 KK852811 KDR15922.1 PYGN01000311 PSN48741.1 GEZM01013087 JAV92761.1 GECZ01006610 JAS63159.1 DS235873 EEB19668.1 KQ971335 EEZ99648.1 GGFM01007598 MBW28349.1 AXCN02000366 ADMH02001494 ETN62317.1 GGFJ01007476 MBW56617.1 GGFK01009665 MBW42986.1 GAMD01001029 JAB00562.1 KQ434972 KZC12743.1 UFQT01003315 SSX34846.1 GL450269 EFN81239.1 AAAB01008960 EAA10924.4 AXCM01000869 NNAY01001263 OXU24566.1 ATLV01021973 KE525326 KFB47034.1 DS231886 EDS43337.1 GAPW01002668 JAC10930.1 JXUM01061030 JXUM01061031 JXUM01061032 KQ562131 KXJ76616.1 CH477673 EAT37431.1 KQ435828 KOX71890.1 GANO01000354 JAB59517.1 GL768539 EFZ11211.1 LBMM01002238 KMQ95087.1 JXUM01139534 JXUM01139535 JXUM01139536 JXUM01139537 JXUM01139538 KQ568916 KXJ68844.1 GFDL01008215 JAV26830.1 KZ288400 PBC26264.1 JR052145 AEY61628.1 KA646346 AFP60975.1 KQ978993 KYN26691.1 ADTU01004412 GAKP01000730 JAC58222.1 GBYB01014066 JAG83833.1 KQ976448 KYM85877.1 GL887605 EGI70650.1 GDAI01000367 JAI17236.1 APCN01001538 GEDC01019177 JAS18121.1 KK107419 QOIP01000003 EZA51079.1 RLU24490.1 GL441149 EFN65100.1 KQ977004 KYN06487.1 GFTR01005106 JAW11320.1 KK854006 PTY09125.1 ACPB03000234 GBHO01036075 GBHO01036005 GBHO01014083 GBHO01011912 JAG07529.1 JAG07599.1 JAG29521.1 JAG31692.1 GDHC01012184 GDHC01007040 JAQ06445.1 JAQ11589.1 GDKW01002843 JAI53752.1 CVRI01000075 CRL08547.1 GBGD01002132 JAC86757.1 GBBI01003344 JAC15368.1 GECL01002559 JAP03565.1 KQ769940 OAD52733.1 KQ982821 KYQ50189.1 LNIX01000001 OXA61667.1 GEDC01002497 JAS34801.1 KQ981276 KYN43590.1 EZ421968 EZ422203 ADD18264.1 ADD18396.1 CCAG010018850 JXJN01014867 GDHF01004694 JAI47620.1 KQ414627 KOC67471.1 GAMC01000238 JAC06318.1 GBXI01010828 JAD03464.1 GAMC01000237 JAC06319.1
BABH01015701 KZ150173 PZC72592.1 NWSH01002101 PCG69195.1 AGBW02013268 OWR43790.1 KQ459593 KPI96752.1 KQ461108 KPJ09051.1 NEVH01024428 PNF17070.1 PNF17069.1 KK852811 KDR15922.1 PYGN01000311 PSN48741.1 GEZM01013087 JAV92761.1 GECZ01006610 JAS63159.1 DS235873 EEB19668.1 KQ971335 EEZ99648.1 GGFM01007598 MBW28349.1 AXCN02000366 ADMH02001494 ETN62317.1 GGFJ01007476 MBW56617.1 GGFK01009665 MBW42986.1 GAMD01001029 JAB00562.1 KQ434972 KZC12743.1 UFQT01003315 SSX34846.1 GL450269 EFN81239.1 AAAB01008960 EAA10924.4 AXCM01000869 NNAY01001263 OXU24566.1 ATLV01021973 KE525326 KFB47034.1 DS231886 EDS43337.1 GAPW01002668 JAC10930.1 JXUM01061030 JXUM01061031 JXUM01061032 KQ562131 KXJ76616.1 CH477673 EAT37431.1 KQ435828 KOX71890.1 GANO01000354 JAB59517.1 GL768539 EFZ11211.1 LBMM01002238 KMQ95087.1 JXUM01139534 JXUM01139535 JXUM01139536 JXUM01139537 JXUM01139538 KQ568916 KXJ68844.1 GFDL01008215 JAV26830.1 KZ288400 PBC26264.1 JR052145 AEY61628.1 KA646346 AFP60975.1 KQ978993 KYN26691.1 ADTU01004412 GAKP01000730 JAC58222.1 GBYB01014066 JAG83833.1 KQ976448 KYM85877.1 GL887605 EGI70650.1 GDAI01000367 JAI17236.1 APCN01001538 GEDC01019177 JAS18121.1 KK107419 QOIP01000003 EZA51079.1 RLU24490.1 GL441149 EFN65100.1 KQ977004 KYN06487.1 GFTR01005106 JAW11320.1 KK854006 PTY09125.1 ACPB03000234 GBHO01036075 GBHO01036005 GBHO01014083 GBHO01011912 JAG07529.1 JAG07599.1 JAG29521.1 JAG31692.1 GDHC01012184 GDHC01007040 JAQ06445.1 JAQ11589.1 GDKW01002843 JAI53752.1 CVRI01000075 CRL08547.1 GBGD01002132 JAC86757.1 GBBI01003344 JAC15368.1 GECL01002559 JAP03565.1 KQ769940 OAD52733.1 KQ982821 KYQ50189.1 LNIX01000001 OXA61667.1 GEDC01002497 JAS34801.1 KQ981276 KYN43590.1 EZ421968 EZ422203 ADD18264.1 ADD18396.1 CCAG010018850 JXJN01014867 GDHF01004694 JAI47620.1 KQ414627 KOC67471.1 GAMC01000238 JAC06318.1 GBXI01010828 JAD03464.1 GAMC01000237 JAC06319.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000235965
+ More
UP000027135 UP000245037 UP000009046 UP000007266 UP000075886 UP000000673 UP000075920 UP000075900 UP000076502 UP000069272 UP000075884 UP000075903 UP000008237 UP000007062 UP000076407 UP000075882 UP000075883 UP000215335 UP000002358 UP000075885 UP000076408 UP000030765 UP000002320 UP000069940 UP000249989 UP000075880 UP000008820 UP000053105 UP000036403 UP000075902 UP000095301 UP000242457 UP000005203 UP000078492 UP000005205 UP000078540 UP000007755 UP000075840 UP000053097 UP000279307 UP000000311 UP000078542 UP000095300 UP000015103 UP000183832 UP000079169 UP000075881 UP000075809 UP000198287 UP000091820 UP000078541 UP000092445 UP000092444 UP000092443 UP000092460 UP000078200 UP000053825
UP000027135 UP000245037 UP000009046 UP000007266 UP000075886 UP000000673 UP000075920 UP000075900 UP000076502 UP000069272 UP000075884 UP000075903 UP000008237 UP000007062 UP000076407 UP000075882 UP000075883 UP000215335 UP000002358 UP000075885 UP000076408 UP000030765 UP000002320 UP000069940 UP000249989 UP000075880 UP000008820 UP000053105 UP000036403 UP000075902 UP000095301 UP000242457 UP000005203 UP000078492 UP000005205 UP000078540 UP000007755 UP000075840 UP000053097 UP000279307 UP000000311 UP000078542 UP000095300 UP000015103 UP000183832 UP000079169 UP000075881 UP000075809 UP000198287 UP000091820 UP000078541 UP000092445 UP000092444 UP000092443 UP000092460 UP000078200 UP000053825
PRIDE
Pfam
PF09177 Syntaxin-6_N
Interpro
Gene 3D
ProteinModelPortal
H9IXQ6
A0A2W1BBA7
A0A2A4JAT7
A0A212EQM0
A0A194PTV9
A0A194QZM9
+ More
A0A2J7PL56 A0A2J7PL85 A0A067R1P7 A0A2P8YWW5 A0A1Y1N4C4 A0A1B6GL30 E0W1T6 D6WIT9 A0A2M3ZIL5 A0A182Q2P4 W5JD93 A0A182WB39 A0A2M4BU71 A0A2M4AQB4 A0A182RF82 T1DTD4 A0A154PLN7 A0A336MX58 A0A182FDN5 A0A182NGU9 A0A182VP55 E2BSU7 Q7Q6W8 A0A182XMY3 A0A182LJ62 A0A182MUH8 A0A232F1B1 K7J6B2 A0A182PHL0 A0A182Y189 A0A084W9Z0 B0WC92 A0A023EPJ3 A0A182GG38 A0A182J0F2 Q16SG8 A0A0M8ZVC9 U5EYE1 E9J7J4 A0A0J7KX45 A0A182G2J5 A0A1Q3FGW5 A0A182TYS6 A0A1I8NDJ5 A0A2A3E442 V9IKI0 A0A088A5R0 T1PC55 A0A151JLL2 A0A158NYZ3 A0A034WV86 A0A0C9S0H5 A0A151I5C6 F4W554 A0A0K8TSX6 A0A182HWT7 A0A1B6CXK6 A0A026W7R2 E2AN98 A0A195D0U7 A0A1I8PG99 A0A224XSA5 A0A2R7VT43 T1IE43 A0A0A9YF54 A0A146LWN8 A0A0N7Z8N6 A0A1J1J823 A0A069DSW4 A0A1S4EGW3 A0A023F1Z5 A0A0V0G6D3 A0A182JV78 A0A310SBX5 A0A151WQL2 A0A226EWY4 A0A1A9WP66 A0A1B6EA46 A0A195FSM5 A0A240SX67 D3TKN6 A0A240SWW5 A0A1A9Y6J6 A0A1B0BIB0 A0A1A9UJ22 A0A0K8W9U2 A0A0L7R9N8 W8C2Z8 A0A0A1WWM4 W8C7Z4
A0A2J7PL56 A0A2J7PL85 A0A067R1P7 A0A2P8YWW5 A0A1Y1N4C4 A0A1B6GL30 E0W1T6 D6WIT9 A0A2M3ZIL5 A0A182Q2P4 W5JD93 A0A182WB39 A0A2M4BU71 A0A2M4AQB4 A0A182RF82 T1DTD4 A0A154PLN7 A0A336MX58 A0A182FDN5 A0A182NGU9 A0A182VP55 E2BSU7 Q7Q6W8 A0A182XMY3 A0A182LJ62 A0A182MUH8 A0A232F1B1 K7J6B2 A0A182PHL0 A0A182Y189 A0A084W9Z0 B0WC92 A0A023EPJ3 A0A182GG38 A0A182J0F2 Q16SG8 A0A0M8ZVC9 U5EYE1 E9J7J4 A0A0J7KX45 A0A182G2J5 A0A1Q3FGW5 A0A182TYS6 A0A1I8NDJ5 A0A2A3E442 V9IKI0 A0A088A5R0 T1PC55 A0A151JLL2 A0A158NYZ3 A0A034WV86 A0A0C9S0H5 A0A151I5C6 F4W554 A0A0K8TSX6 A0A182HWT7 A0A1B6CXK6 A0A026W7R2 E2AN98 A0A195D0U7 A0A1I8PG99 A0A224XSA5 A0A2R7VT43 T1IE43 A0A0A9YF54 A0A146LWN8 A0A0N7Z8N6 A0A1J1J823 A0A069DSW4 A0A1S4EGW3 A0A023F1Z5 A0A0V0G6D3 A0A182JV78 A0A310SBX5 A0A151WQL2 A0A226EWY4 A0A1A9WP66 A0A1B6EA46 A0A195FSM5 A0A240SX67 D3TKN6 A0A240SWW5 A0A1A9Y6J6 A0A1B0BIB0 A0A1A9UJ22 A0A0K8W9U2 A0A0L7R9N8 W8C2Z8 A0A0A1WWM4 W8C7Z4
PDB
2IFU
E-value=2.90305e-26,
Score=292
Ontologies
GO
PANTHER
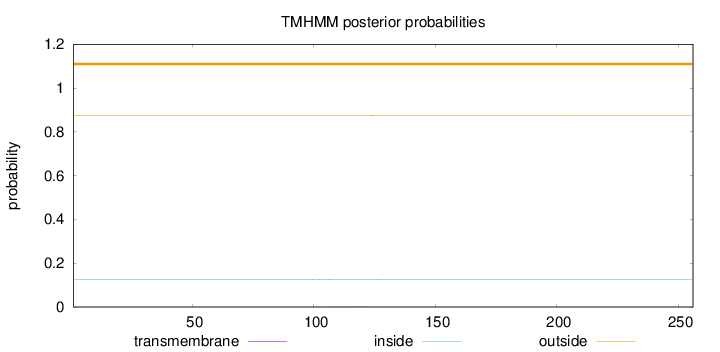
Topology
Length:
256
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03252
Exp number, first 60 AAs:
0.00039
Total prob of N-in:
0.12638
outside
1 - 256
Population Genetic Test Statistics
Pi
22.643858
Theta
20.514208
Tajima's D
-0.68837
CLR
0.00217
CSRT
0.198990050497475
Interpretation
Possibly Positive selection
