Gene
KWMTBOMO00046
Pre Gene Modal
BGIBMGA002040
Annotation
AAEL005572-PA_[Aedes_aegypti]
Full name
ATP-dependent DNA helicase
Location in the cell
Cytoplasmic Reliability : 1.996
Sequence
CDS
ATGGTTTTTAGGGGTTTAAAGGAGAAGATTGACTCTTTGTTTGACCTCGATTTCAAACCAAAAGTACTAGTGAGTGATGCAGCTCCAGCAATTAAAAATGCTTTCTTGGAAGTATTTGGTTTTGATACCACGGTACGCATGTGTTGGGCGCACGCCATAAAAAATATTAAGAAAAAAGTCGAGCAAACGGTGGATAAAAAAAACAGAAAAGACATTATGCAGGATATATATGCTTTACATGACGCATCGTCACAGGAAATTTTCAATGCTGCATCACAAGCTTTTATTGTGAAACATGCTTCAAAGACTTCATTTATAAAATATTTCGAACAAGAATGGCTGATCAAAAATCCTAACTGGTTCTTAGGAGGAGCAGCAACTCCTTCACCGACTACTAACAATGCTTTGGAAGCTTTCAACAAGTCAATCAAAGATCATAATACGATGCGGGAACATTTTCCCTTATCACGATTCCTTACTGTGGCAAGCGAGATGGTTACACATTGGTCTAATGATATCAATGAAAATTCATTCCCAGAAACTCATAATATTGAATTAAAAGAATGGACAAAAGGATATTGTTGGGCAAAACAGAATGTTAAAGTAGACTTTAATATCTGA
Protein
MVFRGLKEKIDSLFDLDFKPKVLVSDAAPAIKNAFLEVFGFDTTVRMCWAHAIKNIKKKVEQTVDKKNRKDIMQDIYALHDASSQEIFNAASQAFIVKHASKTSFIKYFEQEWLIKNPNWFLGGAATPSPTTNNALEAFNKSIKDHNTMREHFPLSRFLTVASEMVTHWSNDINENSFPETHNIELKEWTKGYCWAKQNVKVDFNI
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the helicase family.
Uniprot
H9IXQ7
S4NTK7
A0A1L8DIW7
A0A3M7PNI1
A0A3M7PJM1
H9JJF5
+ More
A0A226D5N1 A0A3M7S999 A0A226DSH1 A0A3M7PAV1 A0A3M7QNK6 A0A3M7PWR2 A0A177BB22 A0A3M7RHF4 A0A226DBB1 A0A226D022 A0A226DU32 A0A226DW34 A0A3M7QAF8 A0A3M7R6I2 A0A3M7T318 A0A226EBP5 A0A177AW62 A0A177B5R4 A0A226DZ44 Q179N8 A0A3M7RZK3 A0A0N8ES31 A0A3M7Q0C3 A0A3M7QS46 A0A3M7QHT4 A0A177AZF6 A0A3M7RYS7 A0A1I7RJ32 A0A226D0L8 A0A3M7SUJ3 A0A3M7Q7J0 A0A3M7QAT9 A0A0W8D799 A0A3M7Q7C0 A0A0W8CSR0 A0A0W8CLG3 W3A7E4 W2XX95 A0A0W8C2M6 A0A0W8CNL5 A0A080YXX4 A0A3M7PHE1 W2VR25 A0A0W8CT04 A0A081A2V5 A0A0W8CEG4 A0A080YWL5 A0A0H5QX69 A0A0W8CJP2 W2XRR1 W2VTQ8 W3A0M6 V9FV77 A0A080ZTY3 W2WKZ5 W2RFQ6 A0A2P4XAL6 A0A147BQB0 W2PMI2 A0A131Y3T8 A0A1W7R615 W2YF10 W2G0D2 W2W403 W2I4E9 C5KSG6 V9EB34 A0A081AYZ8 W2M2G2 W2PEI2 W3A3S7 A0A3M7R9K0 A0A3M7PRH3 W2PFR7 C5L927
A0A226D5N1 A0A3M7S999 A0A226DSH1 A0A3M7PAV1 A0A3M7QNK6 A0A3M7PWR2 A0A177BB22 A0A3M7RHF4 A0A226DBB1 A0A226D022 A0A226DU32 A0A226DW34 A0A3M7QAF8 A0A3M7R6I2 A0A3M7T318 A0A226EBP5 A0A177AW62 A0A177B5R4 A0A226DZ44 Q179N8 A0A3M7RZK3 A0A0N8ES31 A0A3M7Q0C3 A0A3M7QS46 A0A3M7QHT4 A0A177AZF6 A0A3M7RYS7 A0A1I7RJ32 A0A226D0L8 A0A3M7SUJ3 A0A3M7Q7J0 A0A3M7QAT9 A0A0W8D799 A0A3M7Q7C0 A0A0W8CSR0 A0A0W8CLG3 W3A7E4 W2XX95 A0A0W8C2M6 A0A0W8CNL5 A0A080YXX4 A0A3M7PHE1 W2VR25 A0A0W8CT04 A0A081A2V5 A0A0W8CEG4 A0A080YWL5 A0A0H5QX69 A0A0W8CJP2 W2XRR1 W2VTQ8 W3A0M6 V9FV77 A0A080ZTY3 W2WKZ5 W2RFQ6 A0A2P4XAL6 A0A147BQB0 W2PMI2 A0A131Y3T8 A0A1W7R615 W2YF10 W2G0D2 W2W403 W2I4E9 C5KSG6 V9EB34 A0A081AYZ8 W2M2G2 W2PEI2 W3A3S7 A0A3M7R9K0 A0A3M7PRH3 W2PFR7 C5L927
EC Number
3.6.4.12
EMBL
BABH01015693
GAIX01010419
JAA82141.1
GFDF01007676
JAV06408.1
REGN01009642
+ More
RNA00682.1 REGN01010365 RMZ99193.1 BABH01017746 LNIX01000036 OXA39961.1 REGN01001836 RNA32158.1 LNIX01000013 OXA47637.1 REGN01012308 RMZ96099.1 REGN01005530 RNA13007.1 REGN01008621 RNA03118.1 LWCA01000107 OAF70833.1 REGN01003363 RNA23002.1 LNIX01000027 OXA42198.1 LNIX01000047 OXA38214.1 LNIX01000012 OXA48207.1 LNIX01000010 OXA49682.1 REGN01006811 RNA08219.1 REGN01004124 RNA19001.1 REGN01000399 RNA42218.1 LNIX01000005 OXA54574.1 LWCA01000999 OAF66225.1 LWCA01000346 OAF69042.1 LNIX01000009 OXA50320.1 CH477348 EAT42920.1 REGN01002313 RNA28912.1 GDUN01000602 JAN95317.1 REGN01007987 RNA04733.1 REGN01005258 RNA14103.1 REGN01006074 RNA10996.1 LWCA01000709 OAF67250.1 REGN01002381 RNA28508.1 LNIX01000041 OXA39122.1 REGN01000754 RNA39369.1 REGN01007124 RNA07209.1 REGN01006862 RNA08081.1 LNFP01000489 KUF92276.1 REGN01007240 RNA06891.1 LNFP01001245 KUF87046.1 LNFP01001855 KUF84868.1 ANIY01000067 ETP55096.1 ANIX01000071 ETP27147.1 LNFO01005349 KUF78333.1 LNFP01001595 KUF85654.1 ANJA01004234 ETO59235.1 REGN01010785 RMZ98423.1 ANIX01004706 ETP00113.1 LNFP01001191 KUF87278.1 ANJA01001940 ETO73216.1 LNFO01003693 KUF82455.1 ANJA01004790 ETO58776.1 HACM01006146 CRZ06588.1 LNFO01002936 KUF84296.1 ANIX01000381 ETP25182.1 ANIX01004229 ETP01591.1 ANIY01000375 ETP53177.1 ANIZ01000320 ETI55364.1 ANJA01002412 ETO70094.1 ANIX01002627 ETP11210.1 KI669561 ETN24222.1 NCKW01015540 POM62559.1 GEGO01002474 JAR92930.1 KI669622 ETN01806.1 GEFM01002279 JAP73517.1 GEHC01001028 JAV46617.1 ANIY01003767 ETP33387.1 KI688773 ETK75671.1 ANIX01003586 ETP05267.1 KI675568 ETL29099.1 GG676038 EER12580.1 ANIZ01003145 ETI35417.1 ANJA01000361 ETO84109.1 KI696996 ETM30571.1 KI669660 ETM99060.1 ANIY01000374 ETP53184.1 REGN01003882 RNA20293.1 REGN01009229 RNA01660.1 KI669648 ETM99716.1 GG680342 EER06791.1
RNA00682.1 REGN01010365 RMZ99193.1 BABH01017746 LNIX01000036 OXA39961.1 REGN01001836 RNA32158.1 LNIX01000013 OXA47637.1 REGN01012308 RMZ96099.1 REGN01005530 RNA13007.1 REGN01008621 RNA03118.1 LWCA01000107 OAF70833.1 REGN01003363 RNA23002.1 LNIX01000027 OXA42198.1 LNIX01000047 OXA38214.1 LNIX01000012 OXA48207.1 LNIX01000010 OXA49682.1 REGN01006811 RNA08219.1 REGN01004124 RNA19001.1 REGN01000399 RNA42218.1 LNIX01000005 OXA54574.1 LWCA01000999 OAF66225.1 LWCA01000346 OAF69042.1 LNIX01000009 OXA50320.1 CH477348 EAT42920.1 REGN01002313 RNA28912.1 GDUN01000602 JAN95317.1 REGN01007987 RNA04733.1 REGN01005258 RNA14103.1 REGN01006074 RNA10996.1 LWCA01000709 OAF67250.1 REGN01002381 RNA28508.1 LNIX01000041 OXA39122.1 REGN01000754 RNA39369.1 REGN01007124 RNA07209.1 REGN01006862 RNA08081.1 LNFP01000489 KUF92276.1 REGN01007240 RNA06891.1 LNFP01001245 KUF87046.1 LNFP01001855 KUF84868.1 ANIY01000067 ETP55096.1 ANIX01000071 ETP27147.1 LNFO01005349 KUF78333.1 LNFP01001595 KUF85654.1 ANJA01004234 ETO59235.1 REGN01010785 RMZ98423.1 ANIX01004706 ETP00113.1 LNFP01001191 KUF87278.1 ANJA01001940 ETO73216.1 LNFO01003693 KUF82455.1 ANJA01004790 ETO58776.1 HACM01006146 CRZ06588.1 LNFO01002936 KUF84296.1 ANIX01000381 ETP25182.1 ANIX01004229 ETP01591.1 ANIY01000375 ETP53177.1 ANIZ01000320 ETI55364.1 ANJA01002412 ETO70094.1 ANIX01002627 ETP11210.1 KI669561 ETN24222.1 NCKW01015540 POM62559.1 GEGO01002474 JAR92930.1 KI669622 ETN01806.1 GEFM01002279 JAP73517.1 GEHC01001028 JAV46617.1 ANIY01003767 ETP33387.1 KI688773 ETK75671.1 ANIX01003586 ETP05267.1 KI675568 ETL29099.1 GG676038 EER12580.1 ANIZ01003145 ETI35417.1 ANJA01000361 ETO84109.1 KI696996 ETM30571.1 KI669660 ETM99060.1 ANIY01000374 ETP53184.1 REGN01003882 RNA20293.1 REGN01009229 RNA01660.1 KI669648 ETM99716.1 GG680342 EER06791.1
Proteomes
Pfam
Interpro
IPR001207
Transposase_mutator
+ More
IPR018289 MULE_transposase_dom
IPR007527 Znf_SWIM
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR003657 WRKY_dom
IPR036576 WRKY_dom_sf
IPR017956 AT_hook_DNA-bd_motif
IPR038479 Transthyretin-like_sf
IPR025202 PLD-like_dom
IPR011011 Znf_FYVE_PHD
IPR010285 DNA_helicase_pif1-like
IPR013083 Znf_RING/FYVE/PHD
IPR019786 Zinc_finger_PHD-type_CS
IPR027417 P-loop_NTPase
IPR018289 MULE_transposase_dom
IPR007527 Znf_SWIM
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR003657 WRKY_dom
IPR036576 WRKY_dom_sf
IPR017956 AT_hook_DNA-bd_motif
IPR038479 Transthyretin-like_sf
IPR025202 PLD-like_dom
IPR011011 Znf_FYVE_PHD
IPR010285 DNA_helicase_pif1-like
IPR013083 Znf_RING/FYVE/PHD
IPR019786 Zinc_finger_PHD-type_CS
IPR027417 P-loop_NTPase
Gene 3D
ProteinModelPortal
H9IXQ7
S4NTK7
A0A1L8DIW7
A0A3M7PNI1
A0A3M7PJM1
H9JJF5
+ More
A0A226D5N1 A0A3M7S999 A0A226DSH1 A0A3M7PAV1 A0A3M7QNK6 A0A3M7PWR2 A0A177BB22 A0A3M7RHF4 A0A226DBB1 A0A226D022 A0A226DU32 A0A226DW34 A0A3M7QAF8 A0A3M7R6I2 A0A3M7T318 A0A226EBP5 A0A177AW62 A0A177B5R4 A0A226DZ44 Q179N8 A0A3M7RZK3 A0A0N8ES31 A0A3M7Q0C3 A0A3M7QS46 A0A3M7QHT4 A0A177AZF6 A0A3M7RYS7 A0A1I7RJ32 A0A226D0L8 A0A3M7SUJ3 A0A3M7Q7J0 A0A3M7QAT9 A0A0W8D799 A0A3M7Q7C0 A0A0W8CSR0 A0A0W8CLG3 W3A7E4 W2XX95 A0A0W8C2M6 A0A0W8CNL5 A0A080YXX4 A0A3M7PHE1 W2VR25 A0A0W8CT04 A0A081A2V5 A0A0W8CEG4 A0A080YWL5 A0A0H5QX69 A0A0W8CJP2 W2XRR1 W2VTQ8 W3A0M6 V9FV77 A0A080ZTY3 W2WKZ5 W2RFQ6 A0A2P4XAL6 A0A147BQB0 W2PMI2 A0A131Y3T8 A0A1W7R615 W2YF10 W2G0D2 W2W403 W2I4E9 C5KSG6 V9EB34 A0A081AYZ8 W2M2G2 W2PEI2 W3A3S7 A0A3M7R9K0 A0A3M7PRH3 W2PFR7 C5L927
A0A226D5N1 A0A3M7S999 A0A226DSH1 A0A3M7PAV1 A0A3M7QNK6 A0A3M7PWR2 A0A177BB22 A0A3M7RHF4 A0A226DBB1 A0A226D022 A0A226DU32 A0A226DW34 A0A3M7QAF8 A0A3M7R6I2 A0A3M7T318 A0A226EBP5 A0A177AW62 A0A177B5R4 A0A226DZ44 Q179N8 A0A3M7RZK3 A0A0N8ES31 A0A3M7Q0C3 A0A3M7QS46 A0A3M7QHT4 A0A177AZF6 A0A3M7RYS7 A0A1I7RJ32 A0A226D0L8 A0A3M7SUJ3 A0A3M7Q7J0 A0A3M7QAT9 A0A0W8D799 A0A3M7Q7C0 A0A0W8CSR0 A0A0W8CLG3 W3A7E4 W2XX95 A0A0W8C2M6 A0A0W8CNL5 A0A080YXX4 A0A3M7PHE1 W2VR25 A0A0W8CT04 A0A081A2V5 A0A0W8CEG4 A0A080YWL5 A0A0H5QX69 A0A0W8CJP2 W2XRR1 W2VTQ8 W3A0M6 V9FV77 A0A080ZTY3 W2WKZ5 W2RFQ6 A0A2P4XAL6 A0A147BQB0 W2PMI2 A0A131Y3T8 A0A1W7R615 W2YF10 W2G0D2 W2W403 W2I4E9 C5KSG6 V9EB34 A0A081AYZ8 W2M2G2 W2PEI2 W3A3S7 A0A3M7R9K0 A0A3M7PRH3 W2PFR7 C5L927
Ontologies
KEGG
GO
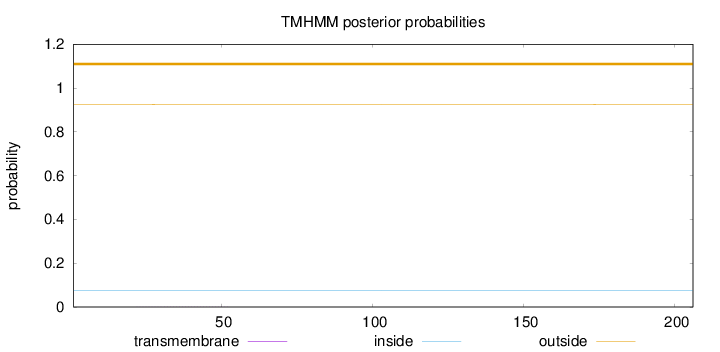
Topology
Length:
206
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01485
Exp number, first 60 AAs:
0.01265
Total prob of N-in:
0.07399
outside
1 - 206
Population Genetic Test Statistics
Pi
20.66984
Theta
22.756017
Tajima's D
-0.261359
CLR
0
CSRT
0.303484825758712
Interpretation
Uncertain
