Gene
KWMTBOMO00040
Pre Gene Modal
BGIBMGA002043
Annotation
PREDICTED:_probable_3'?5'-cyclic_phosphodiesterase_pde-5_isoform_X3_[Amyelois_transitella]
Full name
Phosphodiesterase
+ More
Probable 3',5'-cyclic phosphodiesterase pde-5
Probable 3',5'-cyclic phosphodiesterase pde-5
Location in the cell
Cytoplasmic Reliability : 2.674
Sequence
CDS
ATGTGGAGAACAAAGGACAGGTTCAAAGTCATCGACGAAGAGATTAGTGGTCACATCGTCGTGTGGGGCAGTGTTGCTCTGCATTACTGTAAGCTGTATGTGGATAAGAAAAGGGAACGTAATATGTCCGATTTTTTATTGGACGTTGTCAAAGCAATTTTCGAAGAGATGGTTTCTTTGGATCAACTGATAAAGAGGATATTAGAGTTCGCCCAGAGGCTTGTGAACGCAGACAGGGCTTCGCTATTCTTAGTCGACTACAGGAATTCTGAGCTGGTCTCCACTGTATTCGATCTTAAATTTGAGCCAGGTCTAGATCTGGATATGGAAAAGAAAGAGATTAGGATGCCGATAAATAGAGGAATTGCCGGTCATGTGGCGTTATCAGGCGAGACTATGAATATTCCTGACGCTTATTCTGATTACAGGTTTAATAGAGACGTGGACGAAGCTACTGGATACAAAACAATAAGCATTCTATGTATGCCCATAAAGGTCGAAGGAAAAGTCATTGGCGTTGTCCAAATGGTTAACAAACGCGACGATAAATATTTCGGTGATGACGATGAGGCGGCCTTCGAGATATTCTCCACGTTTTTCGGCCTAGCCCTCCACCACGCGAGGCTTTATGACAGGATCATGAGGAAAGAGCAGAAGTATCGAGTGGCCCTCGAAGTGCTAAGCTATCACAACACGTGCCGGGAATATGAGGTGCAAGCTATGCTGCTAGACAAAGAACCGTTGAATGTTGACTTCGCAGATTTTTATCTAGATCCGTTTCGTTTTGATGAGTTCGAGAAATGTAAATGTATTATATCAATGTTTCGCGATCTGTTTGATATGACCCAGTTTGATATTACGACGCTGACAAGGTTCACGTTGACTGTCAAGAAGAATTATAGGACGGTGCCGTATCATAACTTCGATCATGGCTGGACCGTCGCTCATGCAATGTACGCCATCTTGAAGAGCGATGTGAAGGAGAGATTTGATTATAACAAGAGACTGGCACTCTTCGTTGCGTGTCTTTGCCACGATTTGGACCACCGAGGGTATACCAATAAATACATGAGCGAAACAGCCTCACCTCTAGCCGCCATGTACACAACATCTACATTAGAGCACCATCATTTCAACATTACGGTCACTATTTTACAACAGGACGGTCACAACATATTCTCACATATTTCAAGCGAAGACTACAAAGAGATTCTCGGCTTCATCAGGCACTGTATTCTGGCAACCGATCTAGCGGCCTTCTTCCCTAATTTAGACAAAATAAGAGAATTGTACAGATCTGATGAAGCACAGTTCAGCTGGGACGATCCGAGTCACAGAGATTTGGCCATGGCCATATCAATGACAGCGGCTGATCTGTCAGCTTCAGCTAAGCCATGGGAGATTCAGATCAAAACAGTGAAGGTTATATTTGAGGAGTTTTATGATCAAGGAGACAAGGAGAGAGCTGCTGGAAGGATACCGATACCGATGATGGATAGGAATAAACCTGAAGAGCAGCCAGCTAGCCAGGTTGGTTTCCTCAGTCAAATATGTATACCGTGTTACGACATGCTGTATCGAATACTTCCGAACACAAAGCCGATGTACGCTATGGCAACAAAAAACCTCAACAGATGGAAAGCGAGAGCTGATAAGATCCTTTTTAAGCAAGACTCGAAGAAAGATAATTCTAAAGATAGCCTTGAACCAAAGGATGGAATCGGAGATGATGATGATGATGTCACAGTTAAAAATGAAGGGAACAAAAGAGTAGAAATCATAAGGCAAGAATCAAAAACTATTATAGGAGAAGCTCAACCCAAACCTGTAGTATGGAATGATGACGAAGAATTTGCGGTTGAACAAACCTTTAGAAGTAACATTTGGAAGGAAATTGATGGAGTCACGTGGATCGAAGAAGAAGGCGTATTAGTTGCTAGTGAAGCGACGAAAAAAGATGATACAGTTTGA
Protein
MWRTKDRFKVIDEEISGHIVVWGSVALHYCKLYVDKKRERNMSDFLLDVVKAIFEEMVSLDQLIKRILEFAQRLVNADRASLFLVDYRNSELVSTVFDLKFEPGLDLDMEKKEIRMPINRGIAGHVALSGETMNIPDAYSDYRFNRDVDEATGYKTISILCMPIKVEGKVIGVVQMVNKRDDKYFGDDDEAAFEIFSTFFGLALHHARLYDRIMRKEQKYRVALEVLSYHNTCREYEVQAMLLDKEPLNVDFADFYLDPFRFDEFEKCKCIISMFRDLFDMTQFDITTLTRFTLTVKKNYRTVPYHNFDHGWTVAHAMYAILKSDVKERFDYNKRLALFVACLCHDLDHRGYTNKYMSETASPLAAMYTTSTLEHHHFNITVTILQQDGHNIFSHISSEDYKEILGFIRHCILATDLAAFFPNLDKIRELYRSDEAQFSWDDPSHRDLAMAISMTAADLSASAKPWEIQIKTVKVIFEEFYDQGDKERAAGRIPIPMMDRNKPEEQPASQVGFLSQICIPCYDMLYRILPNTKPMYAMATKNLNRWKARADKILFKQDSKKDNSKDSLEPKDGIGDDDDDVTVKNEGNKRVEIIRQESKTIIGEAQPKPVVWNDDEEFAVEQTFRSNIWKEIDGVTWIEEEGVLVASEATKKDDTV
Summary
Description
Redundantly with pde-1, plays a role in the AFD thermosensory neurons to regulate microvilli receptive ending morphology, possibly by regulating cGMP levels.
Catalytic Activity
a nucleoside 3',5'-cyclic phosphate + H2O = a nucleoside 5'-phosphate + H(+)
Cofactor
a divalent metal cation
Similarity
Belongs to the cyclic nucleotide phosphodiesterase family.
Keywords
cGMP
Coiled coil
Complete proteome
Hydrolase
Metal-binding
Reference proteome
Feature
chain Probable 3',5'-cyclic phosphodiesterase pde-5
Uniprot
H9IXR0
A0A2A4JSA6
A0A2W1C0U8
A0A2H1WFZ1
A0A212EQP0
A0A1W4WYY5
+ More
A0A1W4WN66 A0A1W4WN57 Q16HU8 A0A1S4G0Q2 A0A1B0CY76 F6KPR5 D6WFZ7 A0A0A9XBM5 A0A146LDS5 A0A1J1IKS7 A0A0A9XJX4 A0A0A9X9K5 A0A146KLE9 A0A0A9XBN0 A0A1Q3FNA8 A0A146LFP7 A0A0V0GAE4 A0A067RGF4 B0WPM1 A0A0P4VS35 A0A023F0G6 A0A1B6CTP1 A0A2J7PWX5 A0A0A9YAJ3 A0A0A9XF21 A0A0A9X9L0 A0A0A9XCP1 A0A1B6JSE8 A0A1D2ML08 A0A1B6F059 A0A336LSD4 E9GD11 A0A226EF71 A0A0P4ZJF8 A0A0P6A891 A0A0P5D3J8 A0A164XV76 A0A0P5DQ97 A0A0P5YSN8 A0A0P5T289 A0A0P5CTN9 A0A0P5D9M3 A0A0P5C549 A0A0P5EHR2 A0A0P5J7I7 A0A0P5HR42 A0A069DZ88 A0A0P5KKM9 V5IGH5 A0A0P5CXV1 A0A0P6EXF8 B7QBR4 A0A2R5LG71 A0A1Z5KUZ7 A0A1D1W945 A0A1Y1KVY5 A0A2T7NR97 A0A1S3J205 T2M376 V5GNV3 A0A1W0WNA2 B3RPP8 R7UJM9 A0A3M6U3Q3 T1ID25 A0A210R081 K1RHP0 A0A2B4RJR5 A0A369SEH1 A0A1L8DL06 A7RVE3 A0A0N4ZF26 A0A1I7SAH8 A0A2A2K1L2 A0A0P5FQE5 A0A183BM06 A0A158PMK3 P91119 A0A0D6M523 A0A016WJA0 A0A260YPC4 A0A2P4UZC5 A0A158P8A5 A0A261B1I0 A0A1I7U6X5 A0A0B2UZ40 A0A0K0F5E4 A0A158Q616 A0A158QWI5 A0A0N5BIF8 A0A090LEJ3 A0A2G5VLJ5 A0A0K0DWG2
A0A1W4WN66 A0A1W4WN57 Q16HU8 A0A1S4G0Q2 A0A1B0CY76 F6KPR5 D6WFZ7 A0A0A9XBM5 A0A146LDS5 A0A1J1IKS7 A0A0A9XJX4 A0A0A9X9K5 A0A146KLE9 A0A0A9XBN0 A0A1Q3FNA8 A0A146LFP7 A0A0V0GAE4 A0A067RGF4 B0WPM1 A0A0P4VS35 A0A023F0G6 A0A1B6CTP1 A0A2J7PWX5 A0A0A9YAJ3 A0A0A9XF21 A0A0A9X9L0 A0A0A9XCP1 A0A1B6JSE8 A0A1D2ML08 A0A1B6F059 A0A336LSD4 E9GD11 A0A226EF71 A0A0P4ZJF8 A0A0P6A891 A0A0P5D3J8 A0A164XV76 A0A0P5DQ97 A0A0P5YSN8 A0A0P5T289 A0A0P5CTN9 A0A0P5D9M3 A0A0P5C549 A0A0P5EHR2 A0A0P5J7I7 A0A0P5HR42 A0A069DZ88 A0A0P5KKM9 V5IGH5 A0A0P5CXV1 A0A0P6EXF8 B7QBR4 A0A2R5LG71 A0A1Z5KUZ7 A0A1D1W945 A0A1Y1KVY5 A0A2T7NR97 A0A1S3J205 T2M376 V5GNV3 A0A1W0WNA2 B3RPP8 R7UJM9 A0A3M6U3Q3 T1ID25 A0A210R081 K1RHP0 A0A2B4RJR5 A0A369SEH1 A0A1L8DL06 A7RVE3 A0A0N4ZF26 A0A1I7SAH8 A0A2A2K1L2 A0A0P5FQE5 A0A183BM06 A0A158PMK3 P91119 A0A0D6M523 A0A016WJA0 A0A260YPC4 A0A2P4UZC5 A0A158P8A5 A0A261B1I0 A0A1I7U6X5 A0A0B2UZ40 A0A0K0F5E4 A0A158Q616 A0A158QWI5 A0A0N5BIF8 A0A090LEJ3 A0A2G5VLJ5 A0A0K0DWG2
EC Number
3.1.4.-
3.1.4.17
3.1.4.17
Pubmed
19121390
28756777
22118469
17510324
24964065
18362917
+ More
19820115 25401762 26823975 24845553 27129103 25474469 27289101 21292972 26334808 25765539 28528879 27649274 28004739 24065732 18719581 23254933 30382153 28812685 22992520 30042472 17615350 21909270 9851916 27062922 25730766 26114425 26394399
19820115 25401762 26823975 24845553 27129103 25474469 27289101 21292972 26334808 25765539 28528879 27649274 28004739 24065732 18719581 23254933 30382153 28812685 22992520 30042472 17615350 21909270 9851916 27062922 25730766 26114425 26394399
EMBL
BABH01015688
BABH01015689
NWSH01000779
PCG74290.1
KZ149922
PZC77693.1
+ More
ODYU01008312 SOQ51792.1 AGBW02013268 OWR43795.1 CH478140 EAT33826.1 AJWK01035539 HQ540429 AEG42080.1 KQ971328 EEZ99770.2 GBHO01026195 GBRD01006051 JAG17409.1 JAG59770.1 GDHC01012974 JAQ05655.1 CVRI01000054 CRL00370.1 GBHO01026189 GBRD01016211 JAG17415.1 JAG49615.1 GBHO01026192 JAG17412.1 GDHC01021680 JAP96948.1 GBHO01026190 JAG17414.1 GFDL01005974 JAV29071.1 GDHC01012404 JAQ06225.1 GECL01001192 JAP04932.1 KK852480 KDR22951.1 DS232027 EDS32424.1 GDKW01001345 JAI55250.1 GBBI01004268 JAC14444.1 GEDC01020705 JAS16593.1 NEVH01020867 PNF20838.1 GBHO01014978 GBRD01007277 JAG28626.1 JAG58544.1 GBHO01026191 JAG17413.1 GBHO01026187 GBRD01007276 JAG17417.1 JAG58545.1 GBHO01026193 JAG17411.1 GECU01005597 JAT02110.1 LJIJ01000955 ODM93561.1 GECZ01026288 JAS43481.1 UFQT01000086 SSX19489.1 GL732539 EFX82772.1 LNIX01000004 OXA56283.1 GDIP01213410 JAJ09992.1 GDIP01033240 JAM70475.1 GDIP01163316 JAJ60086.1 LRGB01000944 KZS14597.1 GDIP01169760 JAJ53642.1 GDIP01053884 JAM49831.1 GDIP01132080 JAL71634.1 GDIP01169759 JAJ53643.1 GDIP01175365 JAJ48037.1 GDIP01175366 JAJ48036.1 GDIP01143116 JAJ80286.1 GDIQ01204134 JAK47591.1 GDIQ01226126 JAK25599.1 GBGD01001065 JAC87824.1 GDIQ01208382 JAK43343.1 GANP01006677 JAB77791.1 GDIP01163915 JAJ59487.1 GDIQ01069142 JAN25595.1 ABJB010075929 ABJB010227318 ABJB010487777 ABJB010522530 ABJB010608325 ABJB010685560 ABJB010940632 ABJB011040192 ABJB011094763 ABJB011096231 DS903006 EEC16286.1 GGLE01004289 MBY08415.1 GFJQ02008070 JAV98899.1 BDGG01000019 GAV08828.1 GEZM01075016 JAV64340.1 PZQS01000010 PVD23694.1 HAAD01000279 CDG66511.1 GANP01012413 JAB72055.1 MTYJ01000071 OQV16694.1 DS985242 EDV27675.1 AMQN01007429 KB300677 ELU06420.1 RCHS01002300 RMX48282.1 ACPB03013655 NEDP02001046 OWF54399.1 JH823218 EKC41025.1 LSMT01000474 PFX17416.1 NOWV01000017 RDD45317.1 GFDF01006952 JAV07132.1 DS469543 EDO44626.1 LIAE01009884 PAV67773.1 GDIQ01253576 JAJ98148.1 UYYA01005321 VDM64508.1 FO080144 KE124835 EPB77491.1 JARK01000245 EYC39676.1 NMWX01000912 OZF75035.1 LFJK02001458 POM30403.1 NIPN01000185 OZG04159.1 JPKZ01002921 KHN74347.1 UYYG01000076 VDN52735.1 UYSL01000080 VDL62631.1 LN609529 CEF66568.1 PDUG01000001 PIC52634.1
ODYU01008312 SOQ51792.1 AGBW02013268 OWR43795.1 CH478140 EAT33826.1 AJWK01035539 HQ540429 AEG42080.1 KQ971328 EEZ99770.2 GBHO01026195 GBRD01006051 JAG17409.1 JAG59770.1 GDHC01012974 JAQ05655.1 CVRI01000054 CRL00370.1 GBHO01026189 GBRD01016211 JAG17415.1 JAG49615.1 GBHO01026192 JAG17412.1 GDHC01021680 JAP96948.1 GBHO01026190 JAG17414.1 GFDL01005974 JAV29071.1 GDHC01012404 JAQ06225.1 GECL01001192 JAP04932.1 KK852480 KDR22951.1 DS232027 EDS32424.1 GDKW01001345 JAI55250.1 GBBI01004268 JAC14444.1 GEDC01020705 JAS16593.1 NEVH01020867 PNF20838.1 GBHO01014978 GBRD01007277 JAG28626.1 JAG58544.1 GBHO01026191 JAG17413.1 GBHO01026187 GBRD01007276 JAG17417.1 JAG58545.1 GBHO01026193 JAG17411.1 GECU01005597 JAT02110.1 LJIJ01000955 ODM93561.1 GECZ01026288 JAS43481.1 UFQT01000086 SSX19489.1 GL732539 EFX82772.1 LNIX01000004 OXA56283.1 GDIP01213410 JAJ09992.1 GDIP01033240 JAM70475.1 GDIP01163316 JAJ60086.1 LRGB01000944 KZS14597.1 GDIP01169760 JAJ53642.1 GDIP01053884 JAM49831.1 GDIP01132080 JAL71634.1 GDIP01169759 JAJ53643.1 GDIP01175365 JAJ48037.1 GDIP01175366 JAJ48036.1 GDIP01143116 JAJ80286.1 GDIQ01204134 JAK47591.1 GDIQ01226126 JAK25599.1 GBGD01001065 JAC87824.1 GDIQ01208382 JAK43343.1 GANP01006677 JAB77791.1 GDIP01163915 JAJ59487.1 GDIQ01069142 JAN25595.1 ABJB010075929 ABJB010227318 ABJB010487777 ABJB010522530 ABJB010608325 ABJB010685560 ABJB010940632 ABJB011040192 ABJB011094763 ABJB011096231 DS903006 EEC16286.1 GGLE01004289 MBY08415.1 GFJQ02008070 JAV98899.1 BDGG01000019 GAV08828.1 GEZM01075016 JAV64340.1 PZQS01000010 PVD23694.1 HAAD01000279 CDG66511.1 GANP01012413 JAB72055.1 MTYJ01000071 OQV16694.1 DS985242 EDV27675.1 AMQN01007429 KB300677 ELU06420.1 RCHS01002300 RMX48282.1 ACPB03013655 NEDP02001046 OWF54399.1 JH823218 EKC41025.1 LSMT01000474 PFX17416.1 NOWV01000017 RDD45317.1 GFDF01006952 JAV07132.1 DS469543 EDO44626.1 LIAE01009884 PAV67773.1 GDIQ01253576 JAJ98148.1 UYYA01005321 VDM64508.1 FO080144 KE124835 EPB77491.1 JARK01000245 EYC39676.1 NMWX01000912 OZF75035.1 LFJK02001458 POM30403.1 NIPN01000185 OZG04159.1 JPKZ01002921 KHN74347.1 UYYG01000076 VDN52735.1 UYSL01000080 VDL62631.1 LN609529 CEF66568.1 PDUG01000001 PIC52634.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000192223
UP000008820
UP000092461
+ More
UP000007266 UP000183832 UP000027135 UP000002320 UP000235965 UP000094527 UP000000305 UP000198287 UP000076858 UP000001555 UP000186922 UP000245119 UP000085678 UP000009022 UP000014760 UP000275408 UP000015103 UP000242188 UP000005408 UP000225706 UP000253843 UP000001593 UP000038045 UP000095284 UP000218231 UP000050741 UP000050601 UP000267027 UP000001940 UP000024635 UP000216624 UP000237256 UP000035642 UP000216463 UP000095282 UP000031036 UP000035680 UP000038040 UP000274756 UP000038043 UP000271162 UP000046392 UP000035682 UP000230233 UP000035681
UP000007266 UP000183832 UP000027135 UP000002320 UP000235965 UP000094527 UP000000305 UP000198287 UP000076858 UP000001555 UP000186922 UP000245119 UP000085678 UP000009022 UP000014760 UP000275408 UP000015103 UP000242188 UP000005408 UP000225706 UP000253843 UP000001593 UP000038045 UP000095284 UP000218231 UP000050741 UP000050601 UP000267027 UP000001940 UP000024635 UP000216624 UP000237256 UP000035642 UP000216463 UP000095282 UP000031036 UP000035680 UP000038040 UP000274756 UP000038043 UP000271162 UP000046392 UP000035682 UP000230233 UP000035681
Interpro
IPR002073
PDEase_catalytic_dom
+ More
IPR029016 GAF-like_dom_sf
IPR003607 HD/PDEase_dom
IPR036971 PDEase_catalytic_dom_sf
IPR023174 PDEase_CS
IPR003018 GAF
IPR023088 PDEase
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR021977 Beta-TrCP_D
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR036047 F-box-like_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR029016 GAF-like_dom_sf
IPR003607 HD/PDEase_dom
IPR036971 PDEase_catalytic_dom_sf
IPR023174 PDEase_CS
IPR003018 GAF
IPR023088 PDEase
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR021977 Beta-TrCP_D
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR036047 F-box-like_dom_sf
IPR020472 G-protein_beta_WD-40_rep
Gene 3D
CDD
ProteinModelPortal
H9IXR0
A0A2A4JSA6
A0A2W1C0U8
A0A2H1WFZ1
A0A212EQP0
A0A1W4WYY5
+ More
A0A1W4WN66 A0A1W4WN57 Q16HU8 A0A1S4G0Q2 A0A1B0CY76 F6KPR5 D6WFZ7 A0A0A9XBM5 A0A146LDS5 A0A1J1IKS7 A0A0A9XJX4 A0A0A9X9K5 A0A146KLE9 A0A0A9XBN0 A0A1Q3FNA8 A0A146LFP7 A0A0V0GAE4 A0A067RGF4 B0WPM1 A0A0P4VS35 A0A023F0G6 A0A1B6CTP1 A0A2J7PWX5 A0A0A9YAJ3 A0A0A9XF21 A0A0A9X9L0 A0A0A9XCP1 A0A1B6JSE8 A0A1D2ML08 A0A1B6F059 A0A336LSD4 E9GD11 A0A226EF71 A0A0P4ZJF8 A0A0P6A891 A0A0P5D3J8 A0A164XV76 A0A0P5DQ97 A0A0P5YSN8 A0A0P5T289 A0A0P5CTN9 A0A0P5D9M3 A0A0P5C549 A0A0P5EHR2 A0A0P5J7I7 A0A0P5HR42 A0A069DZ88 A0A0P5KKM9 V5IGH5 A0A0P5CXV1 A0A0P6EXF8 B7QBR4 A0A2R5LG71 A0A1Z5KUZ7 A0A1D1W945 A0A1Y1KVY5 A0A2T7NR97 A0A1S3J205 T2M376 V5GNV3 A0A1W0WNA2 B3RPP8 R7UJM9 A0A3M6U3Q3 T1ID25 A0A210R081 K1RHP0 A0A2B4RJR5 A0A369SEH1 A0A1L8DL06 A7RVE3 A0A0N4ZF26 A0A1I7SAH8 A0A2A2K1L2 A0A0P5FQE5 A0A183BM06 A0A158PMK3 P91119 A0A0D6M523 A0A016WJA0 A0A260YPC4 A0A2P4UZC5 A0A158P8A5 A0A261B1I0 A0A1I7U6X5 A0A0B2UZ40 A0A0K0F5E4 A0A158Q616 A0A158QWI5 A0A0N5BIF8 A0A090LEJ3 A0A2G5VLJ5 A0A0K0DWG2
A0A1W4WN66 A0A1W4WN57 Q16HU8 A0A1S4G0Q2 A0A1B0CY76 F6KPR5 D6WFZ7 A0A0A9XBM5 A0A146LDS5 A0A1J1IKS7 A0A0A9XJX4 A0A0A9X9K5 A0A146KLE9 A0A0A9XBN0 A0A1Q3FNA8 A0A146LFP7 A0A0V0GAE4 A0A067RGF4 B0WPM1 A0A0P4VS35 A0A023F0G6 A0A1B6CTP1 A0A2J7PWX5 A0A0A9YAJ3 A0A0A9XF21 A0A0A9X9L0 A0A0A9XCP1 A0A1B6JSE8 A0A1D2ML08 A0A1B6F059 A0A336LSD4 E9GD11 A0A226EF71 A0A0P4ZJF8 A0A0P6A891 A0A0P5D3J8 A0A164XV76 A0A0P5DQ97 A0A0P5YSN8 A0A0P5T289 A0A0P5CTN9 A0A0P5D9M3 A0A0P5C549 A0A0P5EHR2 A0A0P5J7I7 A0A0P5HR42 A0A069DZ88 A0A0P5KKM9 V5IGH5 A0A0P5CXV1 A0A0P6EXF8 B7QBR4 A0A2R5LG71 A0A1Z5KUZ7 A0A1D1W945 A0A1Y1KVY5 A0A2T7NR97 A0A1S3J205 T2M376 V5GNV3 A0A1W0WNA2 B3RPP8 R7UJM9 A0A3M6U3Q3 T1ID25 A0A210R081 K1RHP0 A0A2B4RJR5 A0A369SEH1 A0A1L8DL06 A7RVE3 A0A0N4ZF26 A0A1I7SAH8 A0A2A2K1L2 A0A0P5FQE5 A0A183BM06 A0A158PMK3 P91119 A0A0D6M523 A0A016WJA0 A0A260YPC4 A0A2P4UZC5 A0A158P8A5 A0A261B1I0 A0A1I7U6X5 A0A0B2UZ40 A0A0K0F5E4 A0A158Q616 A0A158QWI5 A0A0N5BIF8 A0A090LEJ3 A0A2G5VLJ5 A0A0K0DWG2
PDB
3IBJ
E-value=1.71354e-91,
Score=859
Ontologies
GO
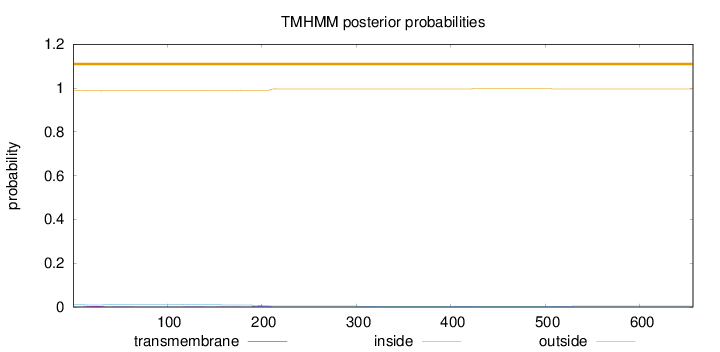
Topology
Length:
656
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.275130000000001
Exp number, first 60 AAs:
0.0819099999999999
Total prob of N-in:
0.01016
outside
1 - 656
Population Genetic Test Statistics
Pi
16.159269
Theta
21.027946
Tajima's D
-0.988573
CLR
0.065179
CSRT
0.138793060346983
Interpretation
Uncertain
