Gene
KWMTBOMO00037
Pre Gene Modal
BGIBMGA002044
Annotation
PREDICTED:_antigen_LPMC-61_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.049 Extracellular Reliability : 1.026 Nuclear Reliability : 1.081
Sequence
CDS
ATGGTCACCGTCAGTGCAGATAGTCACCACCGCTTCAAAATGAGGGTGCTCACTGCTACGATTTGGGTGGTGCTGGCCATCGCGGCATCGGTAGCCGTACCGATCCAGGACGTGTCAAGCGACACCGAGTTAACAAGGGAAAAACGTTCTCCTGGAGGATGGCACTCGCCACCGCCGGAAGGTGTTTGGAAGAAGAAACTCGTCTGGAAGGAAGAGTGGAAGCAGATCTGGAAAACCGAAAAAAAACTCGCCTGGAAATCTGAGTGGAAGAAAGAAAAAGAACCCGCCTGGAAAACTGAACAGGTACAACAGTGGAAAGAAGAACAGGTGCCAGCCTGGAAGGTAGTACAGAAACCTATTTGGAAAGAAGTGCAAGTACCGGTGTGGAAAGAAGTTCAAGTGCCTGATTGGAAGAAAGTCTGGAAACCGGTATGGAAAGAAATACAAGTTCCGGTAACGAAAGACATTCAGGTGCCAGAATGGAAACAGAACTTTATACCTGCTTGGGTCAAAGAGGGCGTGCCCGGTGAGAAATATCTAGGGAAAGATGATCACGGCTGGGAATATACCAGTCACGATCTATGGAGAAAAAAAATGATCTGGAAACCGGTATGGAAGAAAGTTTGGAAGACCGTGCAGAAGCAAGAGTGGGTCACCGAGAAGAAATTGGAATGGAAAGAAGAATGGAAACAAATCTGGAAAACTGAAAAGAAACAGGCTTGGGTAACCGAGAAAAAACAGGAATGGGTAGACGAGAAGGTGCAAATTTGGAAAACCGTTAAGAAAGAAGTCTGGGTGCCGGTACAGAAGAAAATATGGGTTGAGAAATGGAAGCAAATTCAAGTACCGATCTGGAAGACCATAGAAGTGCCCGCATGGAAAAAGGTATGGAAGCCGGTTTGGGAGAAGGTTTGGGTACCGATTCATCATGAGCACCACGAACATCACGGATGGGATTGA
Protein
MVTVSADSHHRFKMRVLTATIWVVLAIAASVAVPIQDVSSDTELTREKRSPGGWHSPPPEGVWKKKLVWKEEWKQIWKTEKKLAWKSEWKKEKEPAWKTEQVQQWKEEQVPAWKVVQKPIWKEVQVPVWKEVQVPDWKKVWKPVWKEIQVPVTKDIQVPEWKQNFIPAWVKEGVPGEKYLGKDDHGWEYTSHDLWRKKMIWKPVWKKVWKTVQKQEWVTEKKLEWKEEWKQIWKTEKKQAWVTEKKQEWVDEKVQIWKTVKKEVWVPVQKKIWVEKWKQIQVPIWKTIEVPAWKKVWKPVWEKVWVPIHHEHHEHHGWD
Summary
Uniprot
H9IXR1
A0A2H1WNQ3
A0A2W1BYD2
A0A2A4JKT5
A0A0L7KV05
I4DLU9
+ More
I4DIB5 A0A212EQP1 A0A194Q2A3 A0A194R1X3 A0A1B0CYN5 E2BXB1 A0A310SGY7 A0A0L7QWY3 E9J2C4 A0A154NYW6 A0A182I0U0 A0A195FXH3 A0A182VSU8 A0A182VCY3 A0A182PK80 A0A182KJV6 A0A2J7R633 A0A158P0R9 A0A182KDH4 T1HVF6 A0A182N8U3 A0A2J7R634 A0A182TYM0 A0A1S4H4E2 A0A1I8Q314 A0A182RAC7 A0A182FYZ1 A0A182JH98 A0A084W5K0 A0A182X7B5 A0A182M288 A0A182TVJ6 A0A1W4WFF6 A0A3B0JD21 U4UJS0 B3MI65 B4KTZ6 Q292T2 B4GCN6 A0A1I8N089 B0W1D8 A0A1W4VE85 B4LN71 N6TKB9 A0A182QQU0 B4HQS9 B4NS40 W8BBH2 H0RNI2 A1Z6X0 B3NA09 Q960I1 A0A182GAC4 B4P1M8 B4J4B7 B4MPZ5 A0A034VYS4 A0A0K8VIM3 J9K1Q3 A0A0A1X3F9 A0A0A1WWZ6 D6WI75 A0A067RMA2 A0A0L0CF31 A0A182T1R2 A0A1S4G7Y8
I4DIB5 A0A212EQP1 A0A194Q2A3 A0A194R1X3 A0A1B0CYN5 E2BXB1 A0A310SGY7 A0A0L7QWY3 E9J2C4 A0A154NYW6 A0A182I0U0 A0A195FXH3 A0A182VSU8 A0A182VCY3 A0A182PK80 A0A182KJV6 A0A2J7R633 A0A158P0R9 A0A182KDH4 T1HVF6 A0A182N8U3 A0A2J7R634 A0A182TYM0 A0A1S4H4E2 A0A1I8Q314 A0A182RAC7 A0A182FYZ1 A0A182JH98 A0A084W5K0 A0A182X7B5 A0A182M288 A0A182TVJ6 A0A1W4WFF6 A0A3B0JD21 U4UJS0 B3MI65 B4KTZ6 Q292T2 B4GCN6 A0A1I8N089 B0W1D8 A0A1W4VE85 B4LN71 N6TKB9 A0A182QQU0 B4HQS9 B4NS40 W8BBH2 H0RNI2 A1Z6X0 B3NA09 Q960I1 A0A182GAC4 B4P1M8 B4J4B7 B4MPZ5 A0A034VYS4 A0A0K8VIM3 J9K1Q3 A0A0A1X3F9 A0A0A1WWZ6 D6WI75 A0A067RMA2 A0A0L0CF31 A0A182T1R2 A0A1S4G7Y8
Pubmed
19121390
28756777
26227816
22651552
22118469
26354079
+ More
20798317 21282665 20966253 21347285 12364791 24438588 23537049 17994087 15632085 25315136 22936249 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 17550304 25348373 25830018 18362917 19820115 24845553 26108605 17510324
20798317 21282665 20966253 21347285 12364791 24438588 23537049 17994087 15632085 25315136 22936249 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 17550304 25348373 25830018 18362917 19820115 24845553 26108605 17510324
EMBL
BABH01015679
ODYU01009906
SOQ54637.1
KZ149922
PZC77696.1
NWSH01001109
+ More
PCG72571.1 JTDY01005537 KOB66906.1 AK402267 BAM18889.1 AK401033 BAM17655.1 AGBW02013268 OWR43797.1 KQ459589 KPI97535.1 KQ460870 KPJ11692.1 AJVK01009261 GL451230 EFN79694.1 KQ767283 OAD53439.1 KQ414705 KOC63105.1 GL767845 EFZ13023.1 KQ434783 KZC04813.1 APCN01005266 KQ981200 KYN45012.1 NEVH01006982 PNF36290.1 ADTU01005771 ACPB03010898 ACPB03010899 ACPB03010900 ACPB03010901 PNF36291.1 AAAB01008980 ATLV01020669 KE525304 KFB45494.1 AXCM01000224 OUUW01000001 SPP73120.1 KB632234 ERL90426.1 CH902619 EDV35910.1 CH933808 EDW08573.2 CM000071 EAL24779.2 CH479181 EDW31484.1 DS231821 EDS45008.1 CH940648 EDW60075.2 APGK01034525 APGK01034526 KB740914 ENN78358.1 AXCN02001866 CH480816 EDW46739.1 CH981838 CM002911 EDX15418.1 KMY91941.1 GAMC01016089 JAB90466.1 BT132904 AEV66554.1 AE013599 AAF59272.1 AHN55955.1 CH954177 EDV59705.1 AY052050 AAK93474.1 JXUM01051008 KQ561671 KXJ77848.1 CM000157 EDW89164.1 CH916367 EDW01599.1 CH963849 EDW74184.1 GAKP01011368 JAC47584.1 GDHF01023449 GDHF01013580 GDHF01001906 JAI28865.1 JAI38734.1 JAI50408.1 ABLF02027206 GBXI01008886 JAD05406.1 GBXI01011354 JAD02938.1 KQ971321 EFA00029.2 KK852542 KDR21720.1 JRES01000487 KNC30822.1
PCG72571.1 JTDY01005537 KOB66906.1 AK402267 BAM18889.1 AK401033 BAM17655.1 AGBW02013268 OWR43797.1 KQ459589 KPI97535.1 KQ460870 KPJ11692.1 AJVK01009261 GL451230 EFN79694.1 KQ767283 OAD53439.1 KQ414705 KOC63105.1 GL767845 EFZ13023.1 KQ434783 KZC04813.1 APCN01005266 KQ981200 KYN45012.1 NEVH01006982 PNF36290.1 ADTU01005771 ACPB03010898 ACPB03010899 ACPB03010900 ACPB03010901 PNF36291.1 AAAB01008980 ATLV01020669 KE525304 KFB45494.1 AXCM01000224 OUUW01000001 SPP73120.1 KB632234 ERL90426.1 CH902619 EDV35910.1 CH933808 EDW08573.2 CM000071 EAL24779.2 CH479181 EDW31484.1 DS231821 EDS45008.1 CH940648 EDW60075.2 APGK01034525 APGK01034526 KB740914 ENN78358.1 AXCN02001866 CH480816 EDW46739.1 CH981838 CM002911 EDX15418.1 KMY91941.1 GAMC01016089 JAB90466.1 BT132904 AEV66554.1 AE013599 AAF59272.1 AHN55955.1 CH954177 EDV59705.1 AY052050 AAK93474.1 JXUM01051008 KQ561671 KXJ77848.1 CM000157 EDW89164.1 CH916367 EDW01599.1 CH963849 EDW74184.1 GAKP01011368 JAC47584.1 GDHF01023449 GDHF01013580 GDHF01001906 JAI28865.1 JAI38734.1 JAI50408.1 ABLF02027206 GBXI01008886 JAD05406.1 GBXI01011354 JAD02938.1 KQ971321 EFA00029.2 KK852542 KDR21720.1 JRES01000487 KNC30822.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053268
UP000053240
+ More
UP000092462 UP000008237 UP000053825 UP000076502 UP000075840 UP000078541 UP000075920 UP000075903 UP000075885 UP000075882 UP000235965 UP000005205 UP000075881 UP000015103 UP000075884 UP000075902 UP000095300 UP000075900 UP000069272 UP000075880 UP000030765 UP000076407 UP000075883 UP000192223 UP000268350 UP000030742 UP000007801 UP000009192 UP000001819 UP000008744 UP000095301 UP000002320 UP000192221 UP000008792 UP000019118 UP000075886 UP000001292 UP000000304 UP000000803 UP000008711 UP000069940 UP000249989 UP000002282 UP000001070 UP000007798 UP000007819 UP000007266 UP000027135 UP000037069 UP000075901
UP000092462 UP000008237 UP000053825 UP000076502 UP000075840 UP000078541 UP000075920 UP000075903 UP000075885 UP000075882 UP000235965 UP000005205 UP000075881 UP000015103 UP000075884 UP000075902 UP000095300 UP000075900 UP000069272 UP000075880 UP000030765 UP000076407 UP000075883 UP000192223 UP000268350 UP000030742 UP000007801 UP000009192 UP000001819 UP000008744 UP000095301 UP000002320 UP000192221 UP000008792 UP000019118 UP000075886 UP000001292 UP000000304 UP000000803 UP000008711 UP000069940 UP000249989 UP000002282 UP000001070 UP000007798 UP000007819 UP000007266 UP000027135 UP000037069 UP000075901
SUPFAM
SSF47240
SSF47240
ProteinModelPortal
H9IXR1
A0A2H1WNQ3
A0A2W1BYD2
A0A2A4JKT5
A0A0L7KV05
I4DLU9
+ More
I4DIB5 A0A212EQP1 A0A194Q2A3 A0A194R1X3 A0A1B0CYN5 E2BXB1 A0A310SGY7 A0A0L7QWY3 E9J2C4 A0A154NYW6 A0A182I0U0 A0A195FXH3 A0A182VSU8 A0A182VCY3 A0A182PK80 A0A182KJV6 A0A2J7R633 A0A158P0R9 A0A182KDH4 T1HVF6 A0A182N8U3 A0A2J7R634 A0A182TYM0 A0A1S4H4E2 A0A1I8Q314 A0A182RAC7 A0A182FYZ1 A0A182JH98 A0A084W5K0 A0A182X7B5 A0A182M288 A0A182TVJ6 A0A1W4WFF6 A0A3B0JD21 U4UJS0 B3MI65 B4KTZ6 Q292T2 B4GCN6 A0A1I8N089 B0W1D8 A0A1W4VE85 B4LN71 N6TKB9 A0A182QQU0 B4HQS9 B4NS40 W8BBH2 H0RNI2 A1Z6X0 B3NA09 Q960I1 A0A182GAC4 B4P1M8 B4J4B7 B4MPZ5 A0A034VYS4 A0A0K8VIM3 J9K1Q3 A0A0A1X3F9 A0A0A1WWZ6 D6WI75 A0A067RMA2 A0A0L0CF31 A0A182T1R2 A0A1S4G7Y8
I4DIB5 A0A212EQP1 A0A194Q2A3 A0A194R1X3 A0A1B0CYN5 E2BXB1 A0A310SGY7 A0A0L7QWY3 E9J2C4 A0A154NYW6 A0A182I0U0 A0A195FXH3 A0A182VSU8 A0A182VCY3 A0A182PK80 A0A182KJV6 A0A2J7R633 A0A158P0R9 A0A182KDH4 T1HVF6 A0A182N8U3 A0A2J7R634 A0A182TYM0 A0A1S4H4E2 A0A1I8Q314 A0A182RAC7 A0A182FYZ1 A0A182JH98 A0A084W5K0 A0A182X7B5 A0A182M288 A0A182TVJ6 A0A1W4WFF6 A0A3B0JD21 U4UJS0 B3MI65 B4KTZ6 Q292T2 B4GCN6 A0A1I8N089 B0W1D8 A0A1W4VE85 B4LN71 N6TKB9 A0A182QQU0 B4HQS9 B4NS40 W8BBH2 H0RNI2 A1Z6X0 B3NA09 Q960I1 A0A182GAC4 B4P1M8 B4J4B7 B4MPZ5 A0A034VYS4 A0A0K8VIM3 J9K1Q3 A0A0A1X3F9 A0A0A1WWZ6 D6WI75 A0A067RMA2 A0A0L0CF31 A0A182T1R2 A0A1S4G7Y8
Ontologies
GO
Topology
SignalP
Position: 1 - 32,
Likelihood: 0.627536
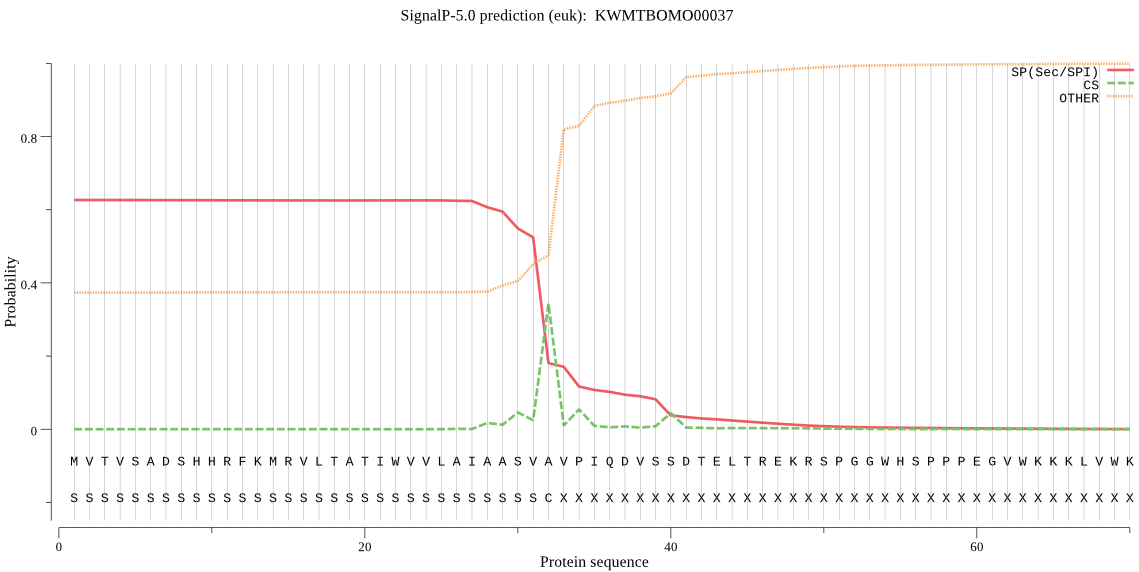
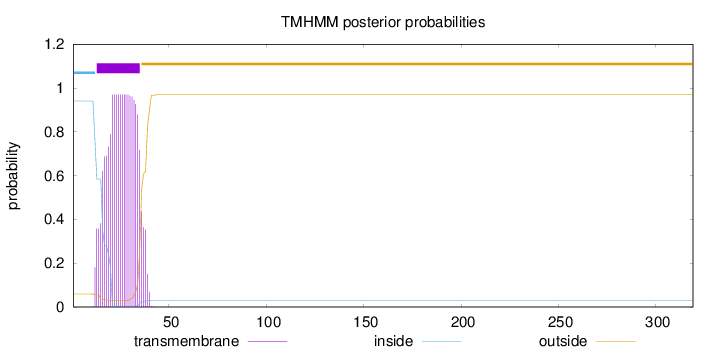
Length:
319
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.30773
Exp number, first 60 AAs:
20.30773
Total prob of N-in:
0.94112
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 319
Population Genetic Test Statistics
Pi
32.813065
Theta
26.440319
Tajima's D
0.992998
CLR
0
CSRT
0.664866756662167
Interpretation
Uncertain
