Gene
KWMTBOMO00033 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002085
Annotation
CYP303A1_[Helicoverpa_armigera]
Full name
Probable cytochrome P450 303a1
+ More
Methyl farnesoate epoxidase
Methyl farnesoate epoxidase
Alternative Name
CYPCCCIIIA1
Cytochrome P450 CYP15A1
Cytochrome P450 CYP15A1
Location in the cell
Cytoplasmic Reliability : 1.403 Mitochondrial Reliability : 1.467
Sequence
CDS
ATGTGGCTGGCAGCGCTAGCGGTTCTGTTAGCGGTGTGCTTGTATCTCTTTTTGGATACCCTCAAACCGAGAAAATTCCCTCCCGGCCCGAAATGGACACCGATTCTTGGCTGCGCAAAGGAAGTTTACAAGCTTCGAGAAAAAACCGGCTACCTGTACAAAGCCGTTAGAGAATTATCTCTGACCTACTGCAAGGAGACTCCAGTACTAGGTTTGAGAATAGGGAAAGACCGAATCGTGATGGTGAATAGCTTGGAAGCAAACAAAGAGATGTTATTCAATGAGGACATCGACGGAAGACCGAAGGGAATCTTTTATCAAACCAGAACGTGGGGAGAACGACGAGGTGTCTTGTTAACAGACGGGGAACTGTGGAAGGAGCAGAGGCGCTTCCTCATCAAGCACCTTAAAGAATTCGGCTTTGGCAGATCCGGTATGGGCGAGACAGCGAAACTGGAAGCCGAGCACATAGTCATCGACGTAATGCACATGATCGGCGACAGGGGATCAGCCGTTATACAAATGCACAACTTCTTCAACGTTTATATTCTGAATACTCTATGGACGATGATGGCCGGGAATAGGTACAATCCAAGCGACCCACAGATGAAGATCCTTCAAAGCATGCTGTTCGATTTGTTCGCAGCCGTCGACATGGTCGGTACAGCCTTCAGTCACTTCCCGATATTGAGCATCGTAGCACCGACCTTATCCGGTTATAGAAATTTCATTAAAACGCATAAGAGAATCTGGAAGTTCTTGAGAGAAGAACTAGCGAGACATAAAGACAGTTTTCAGCCAGATAAAGAGGATAAGGACTTTATGGATGTGTACATAAGGGCTTTGCGTGAGCACGGAGAAGTGAACACGTATTCCGAGGGACAGCTCGTGGCCATGTGCATGGACATGTTCATGGCGGGCACGGAGACGACCAGCAAGAGTATGAGCTTCTGCTTCAGCTATCTCGTTAGAGAGCAGGAAGTCCAAAGGAAAGCTCAGGAGGAAATTGACAGGGTTGTAGGAAAAGATCGGGTACCGAGCGTGAACGATAGGCCTAACATGCCCTATAACGAAGCAATAGTCCATGAGTGTGTCAGACACTTTATGGGACGTACCTTCGGTGTACCGCATCGGGCTCTACGAGACACCACTTTAGCTGGATGCCATATACCTGAGGACACAATGGTAGTCAGTAACTACACGAACATTCTACTTGACGAGAACTACTTCCCGGAGCCGTATTCCTTCAAGCCTGAGAGGTTCCTGGTAAACGGGAGGGTCAGTCTGCCTGACCACTACTTTCCCTTCGGTCTTGCGAAGCATCGCTGCATGGGCGATGCATTAGCCAAGTGCAATATCTTCGTTTTCACTACTACAATGCTGCAGAGGTTCTCGCTGGTGCCTGTACCGGGGGAAGGTCTGCCGTCCTTGGATCATATGGACGGAGCCACTCCTTCAGCAGCGCCGTTTAAGGCTTTAGTTATACCACGAATTTGA
Protein
MWLAALAVLLAVCLYLFLDTLKPRKFPPGPKWTPILGCAKEVYKLREKTGYLYKAVRELSLTYCKETPVLGLRIGKDRIVMVNSLEANKEMLFNEDIDGRPKGIFYQTRTWGERRGVLLTDGELWKEQRRFLIKHLKEFGFGRSGMGETAKLEAEHIVIDVMHMIGDRGSAVIQMHNFFNVYILNTLWTMMAGNRYNPSDPQMKILQSMLFDLFAAVDMVGTAFSHFPILSIVAPTLSGYRNFIKTHKRIWKFLREELARHKDSFQPDKEDKDFMDVYIRALREHGEVNTYSEGQLVAMCMDMFMAGTETTSKSMSFCFSYLVREQEVQRKAQEEIDRVVGKDRVPSVNDRPNMPYNEAIVHECVRHFMGRTFGVPHRALRDTTLAGCHIPEDTMVVSNYTNILLDENYFPEPYSFKPERFLVNGRVSLPDHYFPFGLAKHRCMGDALAKCNIFVFTTTMLQRFSLVPVPGEGLPSLDHMDGATPSAAPFKALVIPRI
Summary
Description
May be involved in the metabolism of insect hormones and in the breakdown of synthetic insecticides.
Catalyzes the conversion of methyl farnesoate to juvenile hormone III acid (methyl (2E,6E)-(10R)-10,11-epoxy-3,7,11-trimethyl-2,6-dodecadienoate) in juvenile hormone biosynthesis.
Catalyzes the conversion of methyl farnesoate to juvenile hormone III acid (methyl (2E,6E)-(10R)-10,11-epoxy-3,7,11-trimethyl-2,6-dodecadienoate) in juvenile hormone biosynthesis.
Catalytic Activity
methyl (2E,6E)-farnesoate + O2 + reduced [NADPH--hemoprotein reductase] = H(+) + H2O + juvenile hormone III + oxidized [NADPH--hemoprotein reductase]
Cofactor
heme
Biophysicochemical Properties
380 uM for methyl (2E,6E)-farnesoate (with purified enzyme)
26 uM for methyl (2E,6E)-farnesoate (with recombinant enzyme expressed in E.coli)
26 uM for methyl (2E,6E)-farnesoate (with recombinant enzyme expressed in E.coli)
Miscellaneous
The enzyme is present in all insects, except in lepidoptera (moths and butterflies), and is specific for methyl farnesoate. Lepidoptera contain the farnesoate epoxidase, which is specific for farnesoate.
Similarity
Belongs to the cytochrome P450 family.
Keywords
Complete proteome
Endoplasmic reticulum
Heme
Iron
Membrane
Metal-binding
Microsome
Monooxygenase
Oxidoreductase
Reference proteome
Glycoprotein
Signal
Feature
chain Probable cytochrome P450 303a1
Uniprot
H9IXV2
A0A0K0TQ03
A0A0K0TQ10
A0A0K0TQ19
A0A2H1VQG3
A0A2A4JH06
+ More
A0A194QUP5 A0A194PVH1 A0A0L7LAF8 A0A067QWR9 A0A2J7R878 E0VZW4 A0A0A9X2T3 A0A146M3L6 A0A2H4BKC1 R4G5L1 A0A182UYS2 B0WPM4 A0A182XH82 A0A182KYW1 A0A182HR04 A0A182JVQ4 A0A2H8TP40 J9JSS1 A0A182PTI7 Q7Q0Y0 A0A2D1GSE0 Q16MY7 A0A182S4N6 A0A1J1IJF4 A0A139WKQ2 A0A1B0D403 A0A1W4WY09 A0A182G047 A0A182U1F1 A0A0K0LB78 A0A195CB21 W5JGC0 A0A0L7QSC6 A0A0L0BZA4 A0A1A9XVT2 A0A1B0ANX1 A0A151X6S6 A0A195EGE3 A0A1B0A3X4 A0A1I8N5X7 A0A1B0FH35 A0A195FFS5 B4LTE1 A0A0M3QTZ6 A0A1Q1NKZ1 B4KI15 K4JN70 A0A1A9UQZ8 A0A3B0K0V2 B4JAC4 A0A1I8PM70 A0A1A9WNZ4 B4GQM5 B4MZZ0 Q29NL9 R9UDV8 C4PAT7 A0A151I3Z7 A0A1W4VJM7 B3N654 B4Q6Z4 B4NXQ1 X2JA13 Q9V399 B4I210 A0A0K8U0Y7 B3MJP5 A0A182YQN3 A0A1Y1KV34 A0A336LKB5 A0A158NT15 A0A182M230 U4UI40 A0A2P8XTM3 E2AZS9 E2AZS8 A0A3Q0IZM7 A0A1W6L1Q3 A0A0K8VKY7 A0A0M9A6L7 N6SXQ1 C7SHT4 A0A2A3E0Q3 Q6R7M4 A0A1Y1L2U8 D6WFY6 A0A0F7R7X8
A0A194QUP5 A0A194PVH1 A0A0L7LAF8 A0A067QWR9 A0A2J7R878 E0VZW4 A0A0A9X2T3 A0A146M3L6 A0A2H4BKC1 R4G5L1 A0A182UYS2 B0WPM4 A0A182XH82 A0A182KYW1 A0A182HR04 A0A182JVQ4 A0A2H8TP40 J9JSS1 A0A182PTI7 Q7Q0Y0 A0A2D1GSE0 Q16MY7 A0A182S4N6 A0A1J1IJF4 A0A139WKQ2 A0A1B0D403 A0A1W4WY09 A0A182G047 A0A182U1F1 A0A0K0LB78 A0A195CB21 W5JGC0 A0A0L7QSC6 A0A0L0BZA4 A0A1A9XVT2 A0A1B0ANX1 A0A151X6S6 A0A195EGE3 A0A1B0A3X4 A0A1I8N5X7 A0A1B0FH35 A0A195FFS5 B4LTE1 A0A0M3QTZ6 A0A1Q1NKZ1 B4KI15 K4JN70 A0A1A9UQZ8 A0A3B0K0V2 B4JAC4 A0A1I8PM70 A0A1A9WNZ4 B4GQM5 B4MZZ0 Q29NL9 R9UDV8 C4PAT7 A0A151I3Z7 A0A1W4VJM7 B3N654 B4Q6Z4 B4NXQ1 X2JA13 Q9V399 B4I210 A0A0K8U0Y7 B3MJP5 A0A182YQN3 A0A1Y1KV34 A0A336LKB5 A0A158NT15 A0A182M230 U4UI40 A0A2P8XTM3 E2AZS9 E2AZS8 A0A3Q0IZM7 A0A1W6L1Q3 A0A0K8VKY7 A0A0M9A6L7 N6SXQ1 C7SHT4 A0A2A3E0Q3 Q6R7M4 A0A1Y1L2U8 D6WFY6 A0A0F7R7X8
EC Number
1.14.-.-
1.14.14.127
1.14.14.127
Pubmed
19121390
26174024
28756777
26354079
26227816
24845553
+ More
20566863 25401762 26823975 20966253 12364791 17510324 18362917 19820115 20920257 23761445 26108605 25315136 17994087 28092127 24324548 15632085 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10471707 25244985 28004739 21347285 23537049 29403074 20798317 15024118 25921675
20566863 25401762 26823975 20966253 12364791 17510324 18362917 19820115 20920257 23761445 26108605 25315136 17994087 28092127 24324548 15632085 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10471707 25244985 28004739 21347285 23537049 29403074 20798317 15024118 25921675
EMBL
BABH01015663
BABH01015664
BABH01015665
BABH01015666
BABH01015667
KR709083
+ More
AKR53659.1 KR709085 AKR53661.1 KR709084 KZ149922 AKR53660.1 PZC77682.1 ODYU01003657 SOQ42702.1 NWSH01001488 PCG71099.1 KQ461108 KPJ09044.1 KQ459593 KPI96759.1 JTDY01002069 KOB72186.1 KK853156 KDR10510.1 NEVH01006723 PNF37031.1 DS235854 EEB18920.1 GBHO01032186 GBHO01032184 JAG11418.1 JAG11420.1 GDHC01004276 JAQ14353.1 KF984041 AHJ59374.1 ACPB03000224 GAHY01000435 JAA77075.1 DS232027 EDS32427.1 APCN01001629 GFXV01003984 MBW15789.1 ABLF02036361 AAAB01008980 EAA13781.4 MF471376 ATN95982.1 CH477845 EAT35703.1 CVRI01000054 CRL00375.1 KQ971328 KYB28395.1 AJVK01011122 AJVK01011123 KM217008 AIW79971.1 KQ978023 KYM98059.1 ADMH02001289 ETN63136.1 KQ414758 KOC61548.1 JRES01001119 KNC25358.1 JXJN01001070 KQ982476 KYQ56062.1 KQ978957 KYN27308.1 CCAG010021907 KQ981636 KYN38874.1 CH940649 EDW63911.1 CP012523 ALC39749.1 KX660712 AQM57051.1 CH933807 EDW11297.1 JX876491 AFU86421.1 OUUW01000006 SPP81470.1 CH916368 EDW03795.1 CH479187 EDW39897.1 CH963920 EDW77925.1 CH379059 EAL34199.2 JX462969 AGN52762.1 FJ907954 ACQ91145.1 KQ976472 KYM83976.1 CH954177 EDV58092.1 CM000361 CM002910 EDX05202.1 KMY90490.1 CM000157 EDW89676.1 AE014134 AHN54491.1 AY138853 CH480820 EDW54567.1 GDHF01031920 JAI20394.1 CH902620 EDV31384.1 GEZM01077579 GEZM01077578 JAV63266.1 UFQT01000004 SSX17171.1 ADTU01025471 ADTU01025472 ADTU01025473 AXCM01000301 KB632366 ERL93649.1 PYGN01001366 PSN35367.1 GL444277 EFN61058.1 EFN61057.1 KX609519 ARN17926.1 GDHF01012791 JAI39523.1 KQ435724 KOX78300.1 APGK01052905 APGK01052906 KB741216 ENN72534.1 FJ358494 ACO87994.1 KZ288470 PBC25343.1 AY509244 GEZM01072722 JAV65427.1 EFA01264.1 AB987827 BAR72977.1
AKR53659.1 KR709085 AKR53661.1 KR709084 KZ149922 AKR53660.1 PZC77682.1 ODYU01003657 SOQ42702.1 NWSH01001488 PCG71099.1 KQ461108 KPJ09044.1 KQ459593 KPI96759.1 JTDY01002069 KOB72186.1 KK853156 KDR10510.1 NEVH01006723 PNF37031.1 DS235854 EEB18920.1 GBHO01032186 GBHO01032184 JAG11418.1 JAG11420.1 GDHC01004276 JAQ14353.1 KF984041 AHJ59374.1 ACPB03000224 GAHY01000435 JAA77075.1 DS232027 EDS32427.1 APCN01001629 GFXV01003984 MBW15789.1 ABLF02036361 AAAB01008980 EAA13781.4 MF471376 ATN95982.1 CH477845 EAT35703.1 CVRI01000054 CRL00375.1 KQ971328 KYB28395.1 AJVK01011122 AJVK01011123 KM217008 AIW79971.1 KQ978023 KYM98059.1 ADMH02001289 ETN63136.1 KQ414758 KOC61548.1 JRES01001119 KNC25358.1 JXJN01001070 KQ982476 KYQ56062.1 KQ978957 KYN27308.1 CCAG010021907 KQ981636 KYN38874.1 CH940649 EDW63911.1 CP012523 ALC39749.1 KX660712 AQM57051.1 CH933807 EDW11297.1 JX876491 AFU86421.1 OUUW01000006 SPP81470.1 CH916368 EDW03795.1 CH479187 EDW39897.1 CH963920 EDW77925.1 CH379059 EAL34199.2 JX462969 AGN52762.1 FJ907954 ACQ91145.1 KQ976472 KYM83976.1 CH954177 EDV58092.1 CM000361 CM002910 EDX05202.1 KMY90490.1 CM000157 EDW89676.1 AE014134 AHN54491.1 AY138853 CH480820 EDW54567.1 GDHF01031920 JAI20394.1 CH902620 EDV31384.1 GEZM01077579 GEZM01077578 JAV63266.1 UFQT01000004 SSX17171.1 ADTU01025471 ADTU01025472 ADTU01025473 AXCM01000301 KB632366 ERL93649.1 PYGN01001366 PSN35367.1 GL444277 EFN61058.1 EFN61057.1 KX609519 ARN17926.1 GDHF01012791 JAI39523.1 KQ435724 KOX78300.1 APGK01052905 APGK01052906 KB741216 ENN72534.1 FJ358494 ACO87994.1 KZ288470 PBC25343.1 AY509244 GEZM01072722 JAV65427.1 EFA01264.1 AB987827 BAR72977.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000027135
+ More
UP000235965 UP000009046 UP000015103 UP000075903 UP000002320 UP000076407 UP000075882 UP000075840 UP000075881 UP000007819 UP000075885 UP000007062 UP000008820 UP000075900 UP000183832 UP000007266 UP000092462 UP000192223 UP000069272 UP000075902 UP000078542 UP000000673 UP000053825 UP000037069 UP000092443 UP000092460 UP000075809 UP000078492 UP000092445 UP000095301 UP000092444 UP000078541 UP000008792 UP000092553 UP000009192 UP000078200 UP000268350 UP000001070 UP000095300 UP000091820 UP000008744 UP000007798 UP000001819 UP000078540 UP000192221 UP000008711 UP000000304 UP000002282 UP000000803 UP000001292 UP000007801 UP000076408 UP000005205 UP000075883 UP000030742 UP000245037 UP000000311 UP000079169 UP000053105 UP000019118 UP000242457
UP000235965 UP000009046 UP000015103 UP000075903 UP000002320 UP000076407 UP000075882 UP000075840 UP000075881 UP000007819 UP000075885 UP000007062 UP000008820 UP000075900 UP000183832 UP000007266 UP000092462 UP000192223 UP000069272 UP000075902 UP000078542 UP000000673 UP000053825 UP000037069 UP000092443 UP000092460 UP000075809 UP000078492 UP000092445 UP000095301 UP000092444 UP000078541 UP000008792 UP000092553 UP000009192 UP000078200 UP000268350 UP000001070 UP000095300 UP000091820 UP000008744 UP000007798 UP000001819 UP000078540 UP000192221 UP000008711 UP000000304 UP000002282 UP000000803 UP000001292 UP000007801 UP000076408 UP000005205 UP000075883 UP000030742 UP000245037 UP000000311 UP000079169 UP000053105 UP000019118 UP000242457
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
H9IXV2
A0A0K0TQ03
A0A0K0TQ10
A0A0K0TQ19
A0A2H1VQG3
A0A2A4JH06
+ More
A0A194QUP5 A0A194PVH1 A0A0L7LAF8 A0A067QWR9 A0A2J7R878 E0VZW4 A0A0A9X2T3 A0A146M3L6 A0A2H4BKC1 R4G5L1 A0A182UYS2 B0WPM4 A0A182XH82 A0A182KYW1 A0A182HR04 A0A182JVQ4 A0A2H8TP40 J9JSS1 A0A182PTI7 Q7Q0Y0 A0A2D1GSE0 Q16MY7 A0A182S4N6 A0A1J1IJF4 A0A139WKQ2 A0A1B0D403 A0A1W4WY09 A0A182G047 A0A182U1F1 A0A0K0LB78 A0A195CB21 W5JGC0 A0A0L7QSC6 A0A0L0BZA4 A0A1A9XVT2 A0A1B0ANX1 A0A151X6S6 A0A195EGE3 A0A1B0A3X4 A0A1I8N5X7 A0A1B0FH35 A0A195FFS5 B4LTE1 A0A0M3QTZ6 A0A1Q1NKZ1 B4KI15 K4JN70 A0A1A9UQZ8 A0A3B0K0V2 B4JAC4 A0A1I8PM70 A0A1A9WNZ4 B4GQM5 B4MZZ0 Q29NL9 R9UDV8 C4PAT7 A0A151I3Z7 A0A1W4VJM7 B3N654 B4Q6Z4 B4NXQ1 X2JA13 Q9V399 B4I210 A0A0K8U0Y7 B3MJP5 A0A182YQN3 A0A1Y1KV34 A0A336LKB5 A0A158NT15 A0A182M230 U4UI40 A0A2P8XTM3 E2AZS9 E2AZS8 A0A3Q0IZM7 A0A1W6L1Q3 A0A0K8VKY7 A0A0M9A6L7 N6SXQ1 C7SHT4 A0A2A3E0Q3 Q6R7M4 A0A1Y1L2U8 D6WFY6 A0A0F7R7X8
A0A194QUP5 A0A194PVH1 A0A0L7LAF8 A0A067QWR9 A0A2J7R878 E0VZW4 A0A0A9X2T3 A0A146M3L6 A0A2H4BKC1 R4G5L1 A0A182UYS2 B0WPM4 A0A182XH82 A0A182KYW1 A0A182HR04 A0A182JVQ4 A0A2H8TP40 J9JSS1 A0A182PTI7 Q7Q0Y0 A0A2D1GSE0 Q16MY7 A0A182S4N6 A0A1J1IJF4 A0A139WKQ2 A0A1B0D403 A0A1W4WY09 A0A182G047 A0A182U1F1 A0A0K0LB78 A0A195CB21 W5JGC0 A0A0L7QSC6 A0A0L0BZA4 A0A1A9XVT2 A0A1B0ANX1 A0A151X6S6 A0A195EGE3 A0A1B0A3X4 A0A1I8N5X7 A0A1B0FH35 A0A195FFS5 B4LTE1 A0A0M3QTZ6 A0A1Q1NKZ1 B4KI15 K4JN70 A0A1A9UQZ8 A0A3B0K0V2 B4JAC4 A0A1I8PM70 A0A1A9WNZ4 B4GQM5 B4MZZ0 Q29NL9 R9UDV8 C4PAT7 A0A151I3Z7 A0A1W4VJM7 B3N654 B4Q6Z4 B4NXQ1 X2JA13 Q9V399 B4I210 A0A0K8U0Y7 B3MJP5 A0A182YQN3 A0A1Y1KV34 A0A336LKB5 A0A158NT15 A0A182M230 U4UI40 A0A2P8XTM3 E2AZS9 E2AZS8 A0A3Q0IZM7 A0A1W6L1Q3 A0A0K8VKY7 A0A0M9A6L7 N6SXQ1 C7SHT4 A0A2A3E0Q3 Q6R7M4 A0A1Y1L2U8 D6WFY6 A0A0F7R7X8
PDB
3T3Z
E-value=1.20377e-47,
Score=480
Ontologies
PATHWAY
GO
Topology
Subcellular location
Endoplasmic reticulum membrane
Microsome membrane
Microsome membrane
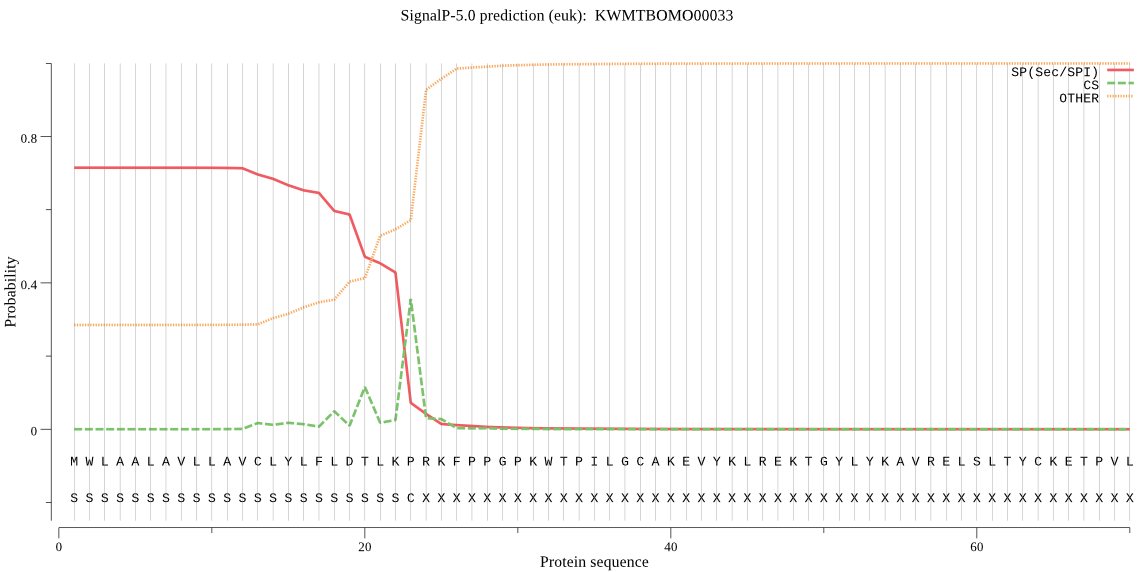
SignalP
Position: 1 - 23,
Likelihood: 0.714576
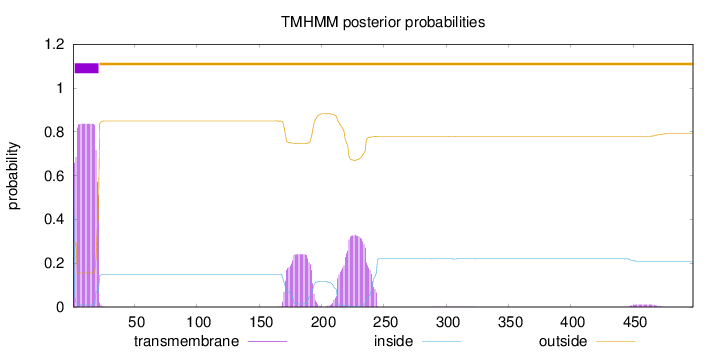
Length:
498
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
28.46438
Exp number, first 60 AAs:
15.69121
Total prob of N-in:
0.70446
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 21
outside
22 - 498
Population Genetic Test Statistics
Pi
4.07242
Theta
6.624764
Tajima's D
-1.01997
CLR
0
CSRT
0.125043747812609
Interpretation
Uncertain
