Gene
KWMTBOMO00030
Pre Gene Modal
BGIBMGA002050
Annotation
PREDICTED:_dipeptidase_1-like_[Bombyx_mori]
Full name
Dipeptidase
Location in the cell
Extracellular Reliability : 1.904
Sequence
CDS
ATGAATCGTTGCGACAGACGTTTGGTGGCATGTGCGCTGTTTGCAATCATCGCGGTGGTTGCAGCCGCTTCCTACGATCGCGAACGGCTGGAGATTGCGAAGCAAATCCTGGAGGAGGTCCCACTCATTGACGGGCATAACGATCTGCCATGGAATATCAGAAAATTCCTTCGGAATCAAGTGAACGACTTCGAACTGGACACAGACCTGACCGTGGTGGAGCCCTGGTCTATCTCCAAGTACTCCCACACTGATCTTCCGAGACTGCGAGAGGGCATGGTGGGAGCTCAAGTGAGTATCGCTAACAATCACGTCTCTGACGCAAAATCATATTTGATGAACGCAGTCAATCTTTTTAAGCCGCTTTTTATAAATAACTAG
Protein
MNRCDRRLVACALFAIIAVVAAASYDRERLEIAKQILEEVPLIDGHNDLPWNIRKFLRNQVNDFELDTDLTVVEPWSISKYSHTDLPRLREGMVGAQVSIANNHVSDAKSYLMNAVNLFKPLFINN
Summary
Catalytic Activity
Hydrolysis of dipeptides.
Cofactor
Zn(2+)
Subunit
Homodimer; disulfide-linked.
Similarity
Belongs to the metallo-dependent hydrolases superfamily. Peptidase M19 family.
Feature
chain Dipeptidase
Uniprot
H9IXR7
A0A2A4JRS2
A0A194QXR7
A0A194PLX8
A0A212EQH0
K7IUE4
+ More
A0A0K8T262 E2BQT2 F4WBI3 A0A1B6BZ16 A0A3L8E3R7 E2AP25 E0VEU5 A0A154PL92 A0A0J7KR01 A0A0A9Y5F3 A0A1B6CMF0 A0A0L7R9C5 E9IBZ0 A0A1A9ULU6 A0A195FVI0 A0A195BHJ7 A0A158P190 A0A182GRQ7 B0WZN2 A0A182FPA6 A0A182JTK7 A0A182VM54 A0A182XAL8 Q7QCH9 A0A182HSF9 A0A182JMV9 A0A182W0S2 A0A182Y6M4 A0A182RQH4 A0A088A5Q6 A0A2A3E3F6 A0A0M8ZY17
A0A0K8T262 E2BQT2 F4WBI3 A0A1B6BZ16 A0A3L8E3R7 E2AP25 E0VEU5 A0A154PL92 A0A0J7KR01 A0A0A9Y5F3 A0A1B6CMF0 A0A0L7R9C5 E9IBZ0 A0A1A9ULU6 A0A195FVI0 A0A195BHJ7 A0A158P190 A0A182GRQ7 B0WZN2 A0A182FPA6 A0A182JTK7 A0A182VM54 A0A182XAL8 Q7QCH9 A0A182HSF9 A0A182JMV9 A0A182W0S2 A0A182Y6M4 A0A182RQH4 A0A088A5Q6 A0A2A3E3F6 A0A0M8ZY17
EC Number
3.4.13.19
Pubmed
EMBL
BABH01015635
NWSH01000802
PCG74110.1
KQ460953
KPJ10333.1
KQ459599
+ More
KPI94332.1 AGBW02013269 OWR43719.1 AAZX01005844 GBRD01006335 JAG59486.1 GL449799 EFN81957.1 GL888063 EGI68462.1 GEDC01031073 JAS06225.1 QOIP01000001 RLU26869.1 GL441439 EFN64857.1 DS235098 EEB11901.1 KQ434958 KZC12626.1 LBMM01004103 KMQ92812.1 GBHO01017276 JAG26328.1 GEDC01022725 JAS14573.1 KQ414627 KOC67480.1 GL762140 EFZ21903.1 KQ981272 KYN43869.1 KQ976488 KYM83641.1 ADTU01006019 ADTU01006020 ADTU01006021 JXUM01082928 JXUM01082929 JXUM01082930 JXUM01082931 KQ563345 KXJ74002.1 DS232211 EDS37546.1 AAAB01008859 EAA08208.5 APCN01000271 KZ288400 PBC26257.1 KQ435828 KOX71899.1
KPI94332.1 AGBW02013269 OWR43719.1 AAZX01005844 GBRD01006335 JAG59486.1 GL449799 EFN81957.1 GL888063 EGI68462.1 GEDC01031073 JAS06225.1 QOIP01000001 RLU26869.1 GL441439 EFN64857.1 DS235098 EEB11901.1 KQ434958 KZC12626.1 LBMM01004103 KMQ92812.1 GBHO01017276 JAG26328.1 GEDC01022725 JAS14573.1 KQ414627 KOC67480.1 GL762140 EFZ21903.1 KQ981272 KYN43869.1 KQ976488 KYM83641.1 ADTU01006019 ADTU01006020 ADTU01006021 JXUM01082928 JXUM01082929 JXUM01082930 JXUM01082931 KQ563345 KXJ74002.1 DS232211 EDS37546.1 AAAB01008859 EAA08208.5 APCN01000271 KZ288400 PBC26257.1 KQ435828 KOX71899.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000002358
+ More
UP000008237 UP000007755 UP000279307 UP000000311 UP000009046 UP000076502 UP000036403 UP000053825 UP000078200 UP000078541 UP000078540 UP000005205 UP000069940 UP000249989 UP000002320 UP000069272 UP000075881 UP000075903 UP000076407 UP000007062 UP000075840 UP000075880 UP000075920 UP000076408 UP000075900 UP000005203 UP000242457 UP000053105
UP000008237 UP000007755 UP000279307 UP000000311 UP000009046 UP000076502 UP000036403 UP000053825 UP000078200 UP000078541 UP000078540 UP000005205 UP000069940 UP000249989 UP000002320 UP000069272 UP000075881 UP000075903 UP000076407 UP000007062 UP000075840 UP000075880 UP000075920 UP000076408 UP000075900 UP000005203 UP000242457 UP000053105
PRIDE
Pfam
PF01244 Peptidase_M19
SUPFAM
SSF51556
SSF51556
CDD
ProteinModelPortal
H9IXR7
A0A2A4JRS2
A0A194QXR7
A0A194PLX8
A0A212EQH0
K7IUE4
+ More
A0A0K8T262 E2BQT2 F4WBI3 A0A1B6BZ16 A0A3L8E3R7 E2AP25 E0VEU5 A0A154PL92 A0A0J7KR01 A0A0A9Y5F3 A0A1B6CMF0 A0A0L7R9C5 E9IBZ0 A0A1A9ULU6 A0A195FVI0 A0A195BHJ7 A0A158P190 A0A182GRQ7 B0WZN2 A0A182FPA6 A0A182JTK7 A0A182VM54 A0A182XAL8 Q7QCH9 A0A182HSF9 A0A182JMV9 A0A182W0S2 A0A182Y6M4 A0A182RQH4 A0A088A5Q6 A0A2A3E3F6 A0A0M8ZY17
A0A0K8T262 E2BQT2 F4WBI3 A0A1B6BZ16 A0A3L8E3R7 E2AP25 E0VEU5 A0A154PL92 A0A0J7KR01 A0A0A9Y5F3 A0A1B6CMF0 A0A0L7R9C5 E9IBZ0 A0A1A9ULU6 A0A195FVI0 A0A195BHJ7 A0A158P190 A0A182GRQ7 B0WZN2 A0A182FPA6 A0A182JTK7 A0A182VM54 A0A182XAL8 Q7QCH9 A0A182HSF9 A0A182JMV9 A0A182W0S2 A0A182Y6M4 A0A182RQH4 A0A088A5Q6 A0A2A3E3F6 A0A0M8ZY17
PDB
3S2N
E-value=7.96747e-10,
Score=146
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
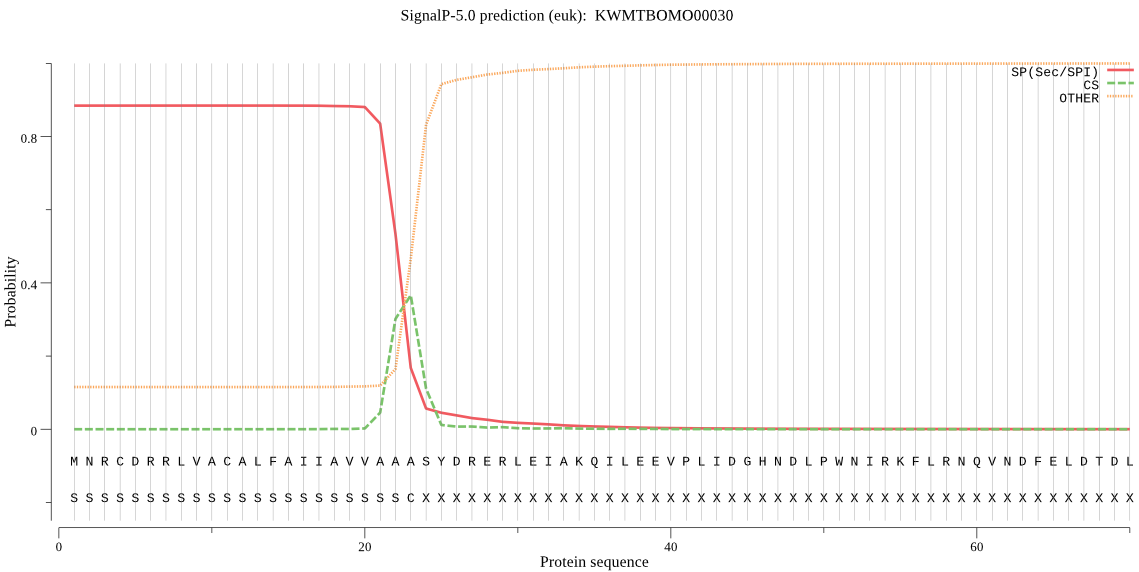
SignalP
Position: 1 - 23,
Likelihood: 0.884624
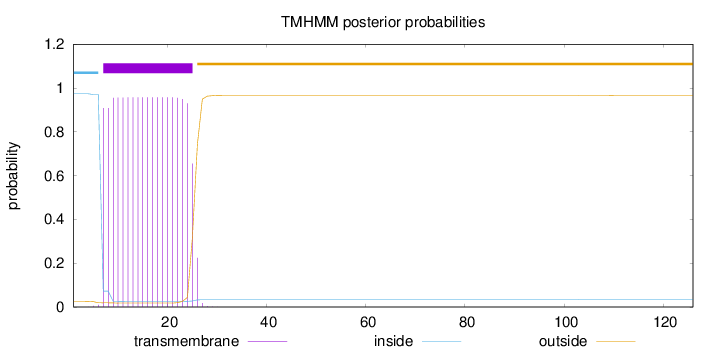
Length:
126
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.01291
Exp number, first 60 AAs:
18.0111
Total prob of N-in:
0.97373
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 25
outside
26 - 126
Population Genetic Test Statistics
Pi
22.009156
Theta
31.677525
Tajima's D
-0.886157
CLR
0.033144
CSRT
0.160541972901355
Interpretation
Uncertain
