Gene
KWMTBOMO00029 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002083
Annotation
beta-actin_[Operophtera_brumata]
Full name
Actin
Location in the cell
Cytoplasmic Reliability : 4.026
Sequence
CDS
ATGTCTTTTGAAAAACCAGCAGTCGTAATAGATAATGGTAGCTATAACATAAAAGCTGGTTTCGCTTGTGACAATCACCCTGTGTCCATATTCAGAACTGTCGTTGGAAGGCCCGACTATCTACACGGGAGCTATGGGAGACAGTCATATGACGTGTATATTGGGGATGACGCGGTCTCCAAGATCGAAGACTTAGAGCTGAACAAGCCAGTGGAGAAAGGCAGAATTGTTCATTGGGATAACATGGAGCGCATCTGGCATCATGTGTTCTATAAGGAGTTGAAGGTGGCACCCGAGGATAGAGCCGTGGTACTGGCCTGTACAACCACATCACCGATGGAGGAAAAGTTCAAATGCTGTGAGGTCTTTTTTGAGACATTAAATGTCCCAGCTTTGTGTATCCAGCCACAGGCCGTTTTAGCCGTCTACGGCTCTGGTTACACGACTGGGATTTCCGTGGACCTTGGATATGATACTACCGAAGTGACTCCAGTATATCAGGGGGGCAGGATTGAGTACGCTCACACTGAAACGCGTTTGGCTGGCTCTCAGGTGTCTGAGTATTTGAAGTTGCAATTAGAAGAACGAGGCATCGATTTAGGTGTGAATACTGCAACGGTTATCGAGAATATTAAAAGGGACTGTCTTTACGTGACCAAAGACGCTTCGATGACTCGAAGTGATTACGAGAAGTACCACAGAACTGCAGATGGAGATATGGTTCACGTTTGTCGGGAAGCGTTCCTGGCTGCAGAATTGATCTTCCAGCCCGATCTGGTAATGGGTCAGAAGACTGATTTCCTTCCTTTGCCGGACGCAGTGGTGAACGCTGTGCTTGTCTGCGATGCGGAGCTGAGACCGGATTTGTATGACGCTATTGTACTTTGTGGTGGATTAGCGATGATTCCAGGTTTACAAGAGAGATTAGCAATGGAAGTGGAGAACCTAACTCAAACACCAGTTAACGTCATAGGATCTCCTGAAGCATACGCCGTAGCTTGGCTTGGCGGCGCCACTTTCGCTGGTCTCCCAGACGCCAAGAAAATGTGGGTCACGAAAAAGCAGTTCGAGGAATATGGCGAGAAAATTGTCAGGAACAAATTCCTATGA
Protein
MSFEKPAVVIDNGSYNIKAGFACDNHPVSIFRTVVGRPDYLHGSYGRQSYDVYIGDDAVSKIEDLELNKPVEKGRIVHWDNMERIWHHVFYKELKVAPEDRAVVLACTTTSPMEEKFKCCEVFFETLNVPALCIQPQAVLAVYGSGYTTGISVDLGYDTTEVTPVYQGGRIEYAHTETRLAGSQVSEYLKLQLEERGIDLGVNTATVIENIKRDCLYVTKDASMTRSDYEKYHRTADGDMVHVCREAFLAAELIFQPDLVMGQKTDFLPLPDAVVNAVLVCDAELRPDLYDAIVLCGGLAMIPGLQERLAMEVENLTQTPVNVIGSPEAYAVAWLGGATFAGLPDAKKMWVTKKQFEEYGEKIVRNKFL
Summary
Similarity
Belongs to the actin family.
Keywords
ATP-binding
Cytoplasm
Cytoskeleton
Nucleotide-binding
Feature
chain Actin
Uniprot
H9IXV0
A0A0L7L7G5
A0A212EQF5
A0A2H1W220
A0A2W1C0W3
A0A2A4JQM4
+ More
R7U3X6 K1QVI5 A0A250WV43 K1RB50 A0A2T9YPQ8 A0A1R1PIQ4 K1RHB1 A0A140JWN7 A0A1E3QP07 A8JAV1 P53498 L1LE80 K1R557 A0A0F7RLL1 A0A2P6NJB2 A0A084RJF5 A0A084AX40 A0A084QBD5 W6KV94 L7I788 G4NGZ4 L7JLM6 A0A250XHD9 A0A0L0NDJ0 A0A061REG3 C5DX74 S6ED84 A0A1S7HQB1 Q2HWE7 C1KGD6 A0A1Y2X5S0 A0A0G4KDZ0 A0A136J705 A0A194W6I0 A0A1W2TLB6 H9C5T9 A0A194VFF0 A0A2T3A5P5 A0A2P5I5A2 A0A168KDA2 E4UZI4 V5N8J4 A0A0P7B081 C5K5X4 A9CAY2 A0A1V8UMK8 A0A1V8S9Z2 O93786 G2Q7Q5 G2QS28 A0A067XMT9 W3X252 A0A0F7RPK1 M7SBZ6 A0A376B355 A0A1V6RAA2 A0A1V6UZ99 K9FUU1 A0A1V6QAV3 A0A0G4MNA7 A0A0G2HAT5 U1G8Q7 A0A167RZ85 B6HG97 A0A1V1SVW0 A0A1Y2VRZ0 A0A3M2SEK2 A0A2H3HM62 A0A2C8CZZ7 A0A2C8D2V7 A0A0W7VGH2 C7YU15 A0A0B0EAU8 A0A2C8CYQ7 A0A395RV34 A0A366RFD7 G9NFJ6 A0A2C8CYX1 A0A2K0WLQ2 A0A2C8D1I6 A0A2C8CZ26 K3W1C1 A0A1Y2UUX3 R8BBG9 S0DZ82 C9S8X4 A0A1L7VGR4 A0A2L2SRB1 A0A2T3ZB76 A0A086ST21 A0A1Y6D5Q7 A0A0M9EXI2 G4UD84 A0A2C8D0F9
R7U3X6 K1QVI5 A0A250WV43 K1RB50 A0A2T9YPQ8 A0A1R1PIQ4 K1RHB1 A0A140JWN7 A0A1E3QP07 A8JAV1 P53498 L1LE80 K1R557 A0A0F7RLL1 A0A2P6NJB2 A0A084RJF5 A0A084AX40 A0A084QBD5 W6KV94 L7I788 G4NGZ4 L7JLM6 A0A250XHD9 A0A0L0NDJ0 A0A061REG3 C5DX74 S6ED84 A0A1S7HQB1 Q2HWE7 C1KGD6 A0A1Y2X5S0 A0A0G4KDZ0 A0A136J705 A0A194W6I0 A0A1W2TLB6 H9C5T9 A0A194VFF0 A0A2T3A5P5 A0A2P5I5A2 A0A168KDA2 E4UZI4 V5N8J4 A0A0P7B081 C5K5X4 A9CAY2 A0A1V8UMK8 A0A1V8S9Z2 O93786 G2Q7Q5 G2QS28 A0A067XMT9 W3X252 A0A0F7RPK1 M7SBZ6 A0A376B355 A0A1V6RAA2 A0A1V6UZ99 K9FUU1 A0A1V6QAV3 A0A0G4MNA7 A0A0G2HAT5 U1G8Q7 A0A167RZ85 B6HG97 A0A1V1SVW0 A0A1Y2VRZ0 A0A3M2SEK2 A0A2H3HM62 A0A2C8CZZ7 A0A2C8D2V7 A0A0W7VGH2 C7YU15 A0A0B0EAU8 A0A2C8CYQ7 A0A395RV34 A0A366RFD7 G9NFJ6 A0A2C8CYX1 A0A2K0WLQ2 A0A2C8D1I6 A0A2C8CZ26 K3W1C1 A0A1Y2UUX3 R8BBG9 S0DZ82 C9S8X4 A0A1L7VGR4 A0A2L2SRB1 A0A2T3ZB76 A0A086ST21 A0A1Y6D5Q7 A0A0M9EXI2 G4UD84 A0A2C8D0F9
Pubmed
19121390
26227816
22118469
28756777
23254933
22992520
+ More
29764946 17932292 8626057 23137308 29378020 25015739 24516393 22876203 15846337 26215153 19525356 23969048 28510588 28078400 27071652 22951933 28684563 10427696 21964414 25623211 27102219 23723393 28368369 23171342 25059858 18820685 19714214 29649280 21501500 23028337 23825955 21829347 28040774 25291769 21750257
29764946 17932292 8626057 23137308 29378020 25015739 24516393 22876203 15846337 26215153 19525356 23969048 28510588 28078400 27071652 22951933 28684563 10427696 21964414 25623211 27102219 23723393 28368369 23171342 25059858 18820685 19714214 29649280 21501500 23028337 23825955 21829347 28040774 25291769 21750257
EMBL
BABH01015635
JTDY01002438
KOB71423.1
AGBW02013269
OWR43716.1
ODYU01005843
+ More
SOQ47131.1 KZ149922 PZC77713.1 NWSH01000802 PCG74109.1 AMQN01010709 KB308450 ELT97855.1 JH816363 EKC40887.1 BEGY01000008 GAX74698.1 EKC40884.1 MBFS01002648 PVU94338.1 LSSK01001067 OMH80837.1 EKC40885.1 LC128310 BAU61514.1 KV454432 ODQ79415.1 DS496151 CM008974 EDO98923.1 PNW74319.1 D50839 D50838 ACOU01000002 EKX73468.1 EKC40883.1 AB973305 BAR79440.1 MDYQ01000071 PRP84019.1 KL657772 KFA76340.1 KL648507 KEY69869.1 KL660862 KFA61270.1 HF955092 CCW64792.1 JH793994 ELQ39113.1 CM001236 EHA47504.1 JH794168 ELQ69137.1 BEGY01000081 GAX82491.1 LFRF01000007 KND92054.1 GBEZ01020305 GBEZ01017279 JAC66356.1 JAC69045.1 CU928178 CAR28385.1 HG316468 CDF91887.1 CP019497 CP019505 AQZ15223.1 AQZ19118.1 AB250390 BDGX01000019 BAE79728.1 GAV49880.1 FJ826616 ACO52877.1 KZ112934 OTB16811.1 CVQH01000225 CRJ83716.1 KQ964248 KXJ92968.1 CM003104 KUI71680.1 DF977454 GAP89099.1 JQ238613 AFD50201.1 KN714822 KUI62608.1 KZ678461 PSR83367.1 MAVT02000253 POS77634.1 AZHF01000001 OAA81549.1 DS989826 EFR03514.1 KF569608 AHA84217.1 LKCW01000312 KPM34678.1 GG672080 GG670791 EER17482.1 EER20135.1 D63852 D63853 BAF91213.1 NAEU01000315 OQO24657.1 NAJO01000075 NAJO01000037 OQN95975.1 OQN99929.1 AB003111 BAA74960.1 CP003003 AEO56914.1 CP003009 AEO63418.1 KC145149 AGO59053.1 KI912113 ETS80213.1 AB973307 LSYV01000098 BAR79442.1 KXZ43235.1 KB707569 EMR61667.1 UFAJ01000088 SSD59081.1 MDYO01000009 OQD98475.1 MDDG01000003 OQE43742.1 AKCT01000182 EKV12302.1 MDYN01000008 OQD86102.1 CVQH01023638 CRK35550.1 LCWF01000039 KKY25710.1 KE720921 ERF73842.1 CM002799 KZN86604.1 AM920435 CAP86492.1 BDMC01000004 GAW12728.1 KZ111630 OTA99217.1 NKUJ01000055 RMJ16001.1 NQOC01000001 PCD40744.1 LT906263 SNU76772.1 LT906237 SNU76748.1 MTYH01000142 PNP37606.1 GG698900 EEU44334.1 KN389643 KHE87345.1 LT905536 SNU76100.1 PXOG01000265 RGP63702.1 QKXC01000160 RBR14955.1 ABDG02000013 EHK50711.1 LT906211 SNU76724.1 MTQA01000054 PNP83203.1 LT906159 SNU76676.1 LT906276 SNU76784.1 AFNW01000083 EKJ75640.1 KZ112555 OTA87661.1 KB933326 EON96641.1 HF679025 CCT66702.1 DS985214 EEY14051.1 FJOF01000004 CZR39462.1 LN649232 CEI39451.1 KZ679260 PTB42059.1 JPKY01000279 KFH40253.1 LT841200 SMH63709.1 JXCE01000087 KPA41722.1 GL891107 EGZ75644.1 LT906185 SNU76700.1
SOQ47131.1 KZ149922 PZC77713.1 NWSH01000802 PCG74109.1 AMQN01010709 KB308450 ELT97855.1 JH816363 EKC40887.1 BEGY01000008 GAX74698.1 EKC40884.1 MBFS01002648 PVU94338.1 LSSK01001067 OMH80837.1 EKC40885.1 LC128310 BAU61514.1 KV454432 ODQ79415.1 DS496151 CM008974 EDO98923.1 PNW74319.1 D50839 D50838 ACOU01000002 EKX73468.1 EKC40883.1 AB973305 BAR79440.1 MDYQ01000071 PRP84019.1 KL657772 KFA76340.1 KL648507 KEY69869.1 KL660862 KFA61270.1 HF955092 CCW64792.1 JH793994 ELQ39113.1 CM001236 EHA47504.1 JH794168 ELQ69137.1 BEGY01000081 GAX82491.1 LFRF01000007 KND92054.1 GBEZ01020305 GBEZ01017279 JAC66356.1 JAC69045.1 CU928178 CAR28385.1 HG316468 CDF91887.1 CP019497 CP019505 AQZ15223.1 AQZ19118.1 AB250390 BDGX01000019 BAE79728.1 GAV49880.1 FJ826616 ACO52877.1 KZ112934 OTB16811.1 CVQH01000225 CRJ83716.1 KQ964248 KXJ92968.1 CM003104 KUI71680.1 DF977454 GAP89099.1 JQ238613 AFD50201.1 KN714822 KUI62608.1 KZ678461 PSR83367.1 MAVT02000253 POS77634.1 AZHF01000001 OAA81549.1 DS989826 EFR03514.1 KF569608 AHA84217.1 LKCW01000312 KPM34678.1 GG672080 GG670791 EER17482.1 EER20135.1 D63852 D63853 BAF91213.1 NAEU01000315 OQO24657.1 NAJO01000075 NAJO01000037 OQN95975.1 OQN99929.1 AB003111 BAA74960.1 CP003003 AEO56914.1 CP003009 AEO63418.1 KC145149 AGO59053.1 KI912113 ETS80213.1 AB973307 LSYV01000098 BAR79442.1 KXZ43235.1 KB707569 EMR61667.1 UFAJ01000088 SSD59081.1 MDYO01000009 OQD98475.1 MDDG01000003 OQE43742.1 AKCT01000182 EKV12302.1 MDYN01000008 OQD86102.1 CVQH01023638 CRK35550.1 LCWF01000039 KKY25710.1 KE720921 ERF73842.1 CM002799 KZN86604.1 AM920435 CAP86492.1 BDMC01000004 GAW12728.1 KZ111630 OTA99217.1 NKUJ01000055 RMJ16001.1 NQOC01000001 PCD40744.1 LT906263 SNU76772.1 LT906237 SNU76748.1 MTYH01000142 PNP37606.1 GG698900 EEU44334.1 KN389643 KHE87345.1 LT905536 SNU76100.1 PXOG01000265 RGP63702.1 QKXC01000160 RBR14955.1 ABDG02000013 EHK50711.1 LT906211 SNU76724.1 MTQA01000054 PNP83203.1 LT906159 SNU76676.1 LT906276 SNU76784.1 AFNW01000083 EKJ75640.1 KZ112555 OTA87661.1 KB933326 EON96641.1 HF679025 CCT66702.1 DS985214 EEY14051.1 FJOF01000004 CZR39462.1 LN649232 CEI39451.1 KZ679260 PTB42059.1 JPKY01000279 KFH40253.1 LT841200 SMH63709.1 JXCE01000087 KPA41722.1 GL891107 EGZ75644.1 LT906185 SNU76700.1
Proteomes
UP000005204
UP000037510
UP000007151
UP000218220
UP000014760
UP000005408
+ More
UP000232323 UP000245609 UP000094336 UP000006906 UP000031512 UP000241769 UP000028540 UP000028045 UP000028524 UP000011086 UP000009058 UP000011085 UP000036947 UP000008536 UP000019375 UP000190375 UP000187013 UP000243732 UP000044602 UP000070501 UP000078559 UP000054516 UP000078576 UP000241462 UP000094444 UP000076881 UP000002669 UP000050424 UP000192386 UP000192596 UP000007322 UP000008181 UP000030651 UP000075714 UP000012174 UP000191612 UP000191500 UP000009882 UP000191672 UP000076449 UP000000724 UP000189293 UP000242955 UP000277212 UP000219517 UP000236546 UP000005206 UP000053122 UP000266234 UP000253153 UP000005426 UP000236664 UP000007978 UP000194361 UP000016800 UP000008698 UP000183971 UP000245910 UP000240493 UP000029964 UP000037904 UP000008513
UP000232323 UP000245609 UP000094336 UP000006906 UP000031512 UP000241769 UP000028540 UP000028045 UP000028524 UP000011086 UP000009058 UP000011085 UP000036947 UP000008536 UP000019375 UP000190375 UP000187013 UP000243732 UP000044602 UP000070501 UP000078559 UP000054516 UP000078576 UP000241462 UP000094444 UP000076881 UP000002669 UP000050424 UP000192386 UP000192596 UP000007322 UP000008181 UP000030651 UP000075714 UP000012174 UP000191612 UP000191500 UP000009882 UP000191672 UP000076449 UP000000724 UP000189293 UP000242955 UP000277212 UP000219517 UP000236546 UP000005206 UP000053122 UP000266234 UP000253153 UP000005426 UP000236664 UP000007978 UP000194361 UP000016800 UP000008698 UP000183971 UP000245910 UP000240493 UP000029964 UP000037904 UP000008513
Pfam
PF00022 Actin
ProteinModelPortal
H9IXV0
A0A0L7L7G5
A0A212EQF5
A0A2H1W220
A0A2W1C0W3
A0A2A4JQM4
+ More
R7U3X6 K1QVI5 A0A250WV43 K1RB50 A0A2T9YPQ8 A0A1R1PIQ4 K1RHB1 A0A140JWN7 A0A1E3QP07 A8JAV1 P53498 L1LE80 K1R557 A0A0F7RLL1 A0A2P6NJB2 A0A084RJF5 A0A084AX40 A0A084QBD5 W6KV94 L7I788 G4NGZ4 L7JLM6 A0A250XHD9 A0A0L0NDJ0 A0A061REG3 C5DX74 S6ED84 A0A1S7HQB1 Q2HWE7 C1KGD6 A0A1Y2X5S0 A0A0G4KDZ0 A0A136J705 A0A194W6I0 A0A1W2TLB6 H9C5T9 A0A194VFF0 A0A2T3A5P5 A0A2P5I5A2 A0A168KDA2 E4UZI4 V5N8J4 A0A0P7B081 C5K5X4 A9CAY2 A0A1V8UMK8 A0A1V8S9Z2 O93786 G2Q7Q5 G2QS28 A0A067XMT9 W3X252 A0A0F7RPK1 M7SBZ6 A0A376B355 A0A1V6RAA2 A0A1V6UZ99 K9FUU1 A0A1V6QAV3 A0A0G4MNA7 A0A0G2HAT5 U1G8Q7 A0A167RZ85 B6HG97 A0A1V1SVW0 A0A1Y2VRZ0 A0A3M2SEK2 A0A2H3HM62 A0A2C8CZZ7 A0A2C8D2V7 A0A0W7VGH2 C7YU15 A0A0B0EAU8 A0A2C8CYQ7 A0A395RV34 A0A366RFD7 G9NFJ6 A0A2C8CYX1 A0A2K0WLQ2 A0A2C8D1I6 A0A2C8CZ26 K3W1C1 A0A1Y2UUX3 R8BBG9 S0DZ82 C9S8X4 A0A1L7VGR4 A0A2L2SRB1 A0A2T3ZB76 A0A086ST21 A0A1Y6D5Q7 A0A0M9EXI2 G4UD84 A0A2C8D0F9
R7U3X6 K1QVI5 A0A250WV43 K1RB50 A0A2T9YPQ8 A0A1R1PIQ4 K1RHB1 A0A140JWN7 A0A1E3QP07 A8JAV1 P53498 L1LE80 K1R557 A0A0F7RLL1 A0A2P6NJB2 A0A084RJF5 A0A084AX40 A0A084QBD5 W6KV94 L7I788 G4NGZ4 L7JLM6 A0A250XHD9 A0A0L0NDJ0 A0A061REG3 C5DX74 S6ED84 A0A1S7HQB1 Q2HWE7 C1KGD6 A0A1Y2X5S0 A0A0G4KDZ0 A0A136J705 A0A194W6I0 A0A1W2TLB6 H9C5T9 A0A194VFF0 A0A2T3A5P5 A0A2P5I5A2 A0A168KDA2 E4UZI4 V5N8J4 A0A0P7B081 C5K5X4 A9CAY2 A0A1V8UMK8 A0A1V8S9Z2 O93786 G2Q7Q5 G2QS28 A0A067XMT9 W3X252 A0A0F7RPK1 M7SBZ6 A0A376B355 A0A1V6RAA2 A0A1V6UZ99 K9FUU1 A0A1V6QAV3 A0A0G4MNA7 A0A0G2HAT5 U1G8Q7 A0A167RZ85 B6HG97 A0A1V1SVW0 A0A1Y2VRZ0 A0A3M2SEK2 A0A2H3HM62 A0A2C8CZZ7 A0A2C8D2V7 A0A0W7VGH2 C7YU15 A0A0B0EAU8 A0A2C8CYQ7 A0A395RV34 A0A366RFD7 G9NFJ6 A0A2C8CYX1 A0A2K0WLQ2 A0A2C8D1I6 A0A2C8CZ26 K3W1C1 A0A1Y2UUX3 R8BBG9 S0DZ82 C9S8X4 A0A1L7VGR4 A0A2L2SRB1 A0A2T3ZB76 A0A086ST21 A0A1Y6D5Q7 A0A0M9EXI2 G4UD84 A0A2C8D0F9
PDB
1YVN
E-value=9.31458e-66,
Score=635
Ontologies
GO
PANTHER
Topology
Subcellular location
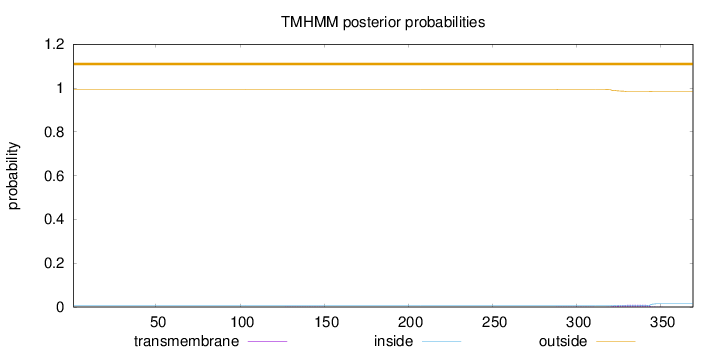
Length:
369
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.21952
Exp number, first 60 AAs:
0.0006
Total prob of N-in:
0.00687
outside
1 - 369
Population Genetic Test Statistics
Pi
25.816099
Theta
26.818852
Tajima's D
-0.510399
CLR
0.01089
CSRT
0.244887755612219
Interpretation
Uncertain
