Gene
KWMTBOMO00028
Pre Gene Modal
BGIBMGA002082
Annotation
actin_[Operophtera_brumata]
Full name
Actin-2
+ More
Actin-7
Actin-7
Alternative Name
Actin-2
Location in the cell
Cytoplasmic Reliability : 2.56
Sequence
CDS
ATGTCAGGCGACGCTATCGGCATTGTGTTGGATATTGGATCCAAATATACTAAGGCTGGTTTCGCAGGAGAATGTTTCCCAAGAGCAGTCTTTCCGAGCACCGTCGGGAGATTTCGTCAATCAGGTCTCGTTGACGGATTCCCAGAGGTCTACTGTGGAAATGAAGCTATTCAAAAAAGAGGAATTTCTCACCTCACTTGGCCGGTAAGCAAAGGGCTGATTCAAGATTGGGATGAGATGGAGAAGCTTTGGCATCACATTTTCTATAAAGGCCTACTTATACCTCCAGAAGCTTGCAGTGTCGTGCACTCGATCCATCCTTTATGCCCAAGGAAGCACCAAGAAAAAATAACAGAAATACTCTTCGAATCATTCACAATCAACTCTCTGTACTTGGCAAGAATGCAGGCTTTAGCTCTGTTTGCAAGCGGTCGGACTACGGGAGTTGTTTGGGAGAGCGGGTACTCATGCTCATACGCAGCCCCAGTCTTTGAAGGATTTCCATTAAGGAATGCCACGTTTATGTCACCGGTCGACGGAGAAACTTTATCTGGACATTTACAAAAATTACTGGCGACTGTAGGCTATTCTTTCACAACTAAAGCGGATAATGACATTTTGGATCAAATTAAGTCTGATAAATGCTACGTGGCGTTAGACTACGATGATGAAGTGGAATCGGGAAAGTGCTCTGGAGATGATAAACACGTTTACACGTTACCGGATGGACAGGAAATATTTTTAGGGCCAGAACGTTTTCAATGTCCAGAGTTGATGTTCCGGCCGAATCTGGGTGGGCTCCAATGTCCGAGTTTGGTGGATTTGTTATGCAAAAGTATCGAGAATTGTGATTTGGATTACCAGCGTGCTTTCTACGAGAATGTTGTGATTGCTGGTGGATCCACTATGTTTCCTGGAATTGTGGATCGGCTACAATTAGAACTGACGAAGAAGTTGTTGAAACGCTCGGAGAATTGGAAGGTTCTCGTCGACGCCATGACAACTAGGCAGTTTGCTGTATGGAACGGTGGATCCATTATGGCCTCTCTGAGTACTATGAACGGTTTTTTGTTGAGTAAAGAAGAATACGAAGATAACGGTTCTGACAGAGTCAAGCATCTATTTTTTTGA
Protein
MSGDAIGIVLDIGSKYTKAGFAGECFPRAVFPSTVGRFRQSGLVDGFPEVYCGNEAIQKRGISHLTWPVSKGLIQDWDEMEKLWHHIFYKGLLIPPEACSVVHSIHPLCPRKHQEKITEILFESFTINSLYLARMQALALFASGRTTGVVWESGYSCSYAAPVFEGFPLRNATFMSPVDGETLSGHLQKLLATVGYSFTTKADNDILDQIKSDKCYVALDYDDEVESGKCSGDDKHVYTLPDGQEIFLGPERFQCPELMFRPNLGGLQCPSLVDLLCKSIENCDLDYQRAFYENVVIAGGSTMFPGIVDRLQLELTKKLLKRSENWKVLVDAMTTRQFAVWNGGSIMASLSTMNGFLLSKEEYEDNGSDRVKHLFF
Summary
Description
Actins are highly conserved proteins that are involved in various types of cell motility and are ubiquitously expressed in all eukaryotic cells. Essential component of cell cytoskeleton; plays an important role in cytoplasmic streaming, cell shape determination, cell division, organelle movement and extension growth. This is considered as one of the vegetative actins which is involved in the regulation of hormone-induced plant cell proliferation and callus formation.
Subunit
Polymerization of globular actin (G-actin) leads to a structural filament (F-actin) in the form of a two-stranded helix. The binding of profilin to monomeric G-actin cause the sequestration of actin into profilactin complexes, and prevents the polymerization.
Miscellaneous
There are at least eight actin genes in rice.
There are 8 actin genes in A.thaliana.
There are 8 actin genes in A.thaliana.
Similarity
Belongs to the actin family.
Keywords
ATP-binding
Complete proteome
Cytoplasm
Cytoskeleton
Nucleotide-binding
Reference proteome
Acetylation
Feature
chain Actin-2
Uniprot
H9IXU9
A0A0L7L7V8
A0A2A4JZW6
A0A2W1BVQ8
A0A2H1W2S8
A0A212EQF7
+ More
M2W5E2 A0A0D9XLE8 A0A0D3HFI5 A0A0E0H4X6 A0A0E0EZS7 A0A0E0R185 A0A0E0BCK4 A0A0E0M9L0 A3C6D7 P0C539 A0A1R3K6S2 A0A1R3JK78 A0A2S3IKW4 A0A2T7C4P4 J3N402 A0A3L6TQH0 A0A3L6S6Y4 B6SI11 A0A059CIZ9 W5AH12 M8BZM2 F2CSU3 M8ASF1 A0A067YNJ5 I1I5C8 A0A0D2MXJ8 A0A2P5RXK1 A0A0B0P7F0 A0A1U8NDN5 M0TX26 A0A0D3CS69 A0A3P6G7D2 C0HHR4 A0A061G2I3 A0A075AJN4 A0A183LCR5 C5WW94 A0A1E5UXJ2 A0A0A7LUI4 G3KF67 A0A200PLG9 A0A2S3HMU2 A0A2U7N5E0 A0A2T7DBC6 A0A1E5VH52 A0A3L6PSP8 K3XQU0 D3JTC9 A0A200PMK0 A0A1U8JZ36 A0A385HDR9 A0A183K206 A0A2P5F5I2 A0A075A1W9 A0A2N9EGA0 A0A0A7LWK3 B4FRH8 A0A2H3Z634 A0A0A9D1Q4 A0A2S3HBM7 A0A2T7ED87 A0A3L6RE42 A0A2T9Z0G9 C5Y567 A0A0E3VJX4 V5YU14 M0TQW8 M0SBB4 A0A2P5AVT0 A0A2T9YPQ8 K7N8I1 K7N907 B4XAN5 E9P1J6 A0A1D6LI44 C5YYR1 D7M276 A0A178UEW4 P53492 H9B8D9 A0A2H3XM37 Q5J1K2 A0A1B3IJS2 A0A218WC81 Q5XPX5 C5YY31 B4F989 A0A0D3HIK8 B8BJ82 A0A0E0MCT4 J3N6A3 A0A0E0F2V9 A0A0D9XPE5 Q67G20
M2W5E2 A0A0D9XLE8 A0A0D3HFI5 A0A0E0H4X6 A0A0E0EZS7 A0A0E0R185 A0A0E0BCK4 A0A0E0M9L0 A3C6D7 P0C539 A0A1R3K6S2 A0A1R3JK78 A0A2S3IKW4 A0A2T7C4P4 J3N402 A0A3L6TQH0 A0A3L6S6Y4 B6SI11 A0A059CIZ9 W5AH12 M8BZM2 F2CSU3 M8ASF1 A0A067YNJ5 I1I5C8 A0A0D2MXJ8 A0A2P5RXK1 A0A0B0P7F0 A0A1U8NDN5 M0TX26 A0A0D3CS69 A0A3P6G7D2 C0HHR4 A0A061G2I3 A0A075AJN4 A0A183LCR5 C5WW94 A0A1E5UXJ2 A0A0A7LUI4 G3KF67 A0A200PLG9 A0A2S3HMU2 A0A2U7N5E0 A0A2T7DBC6 A0A1E5VH52 A0A3L6PSP8 K3XQU0 D3JTC9 A0A200PMK0 A0A1U8JZ36 A0A385HDR9 A0A183K206 A0A2P5F5I2 A0A075A1W9 A0A2N9EGA0 A0A0A7LWK3 B4FRH8 A0A2H3Z634 A0A0A9D1Q4 A0A2S3HBM7 A0A2T7ED87 A0A3L6RE42 A0A2T9Z0G9 C5Y567 A0A0E3VJX4 V5YU14 M0TQW8 M0SBB4 A0A2P5AVT0 A0A2T9YPQ8 K7N8I1 K7N907 B4XAN5 E9P1J6 A0A1D6LI44 C5YYR1 D7M276 A0A178UEW4 P53492 H9B8D9 A0A2H3XM37 Q5J1K2 A0A1B3IJS2 A0A218WC81 Q5XPX5 C5YY31 B4F989 A0A0D3HIK8 B8BJ82 A0A0E0MCT4 J3N6A3 A0A0E0F2V9 A0A0D9XPE5 Q67G20
Pubmed
19121390
26227816
28756777
22118469
23471408
22408737
+ More
12791992 16100779 18089549 24280374 15685292 12869764 2102841 2103449 23481403 30683860 18937034 19965430 24919147 21415278 23075845 23535596 20148030 23257886 25893780 22801500 24916971 19936069 23731509 19189423 28552780 22580951 30157377 23917264 26217707 29764946 26163025 24929230 21478890 27354520 8754679 10048488 27862469 14593172 8852856 11449050 19245862 18507865 16188032 16381971 23299411
12791992 16100779 18089549 24280374 15685292 12869764 2102841 2103449 23481403 30683860 18937034 19965430 24919147 21415278 23075845 23535596 20148030 23257886 25893780 22801500 24916971 19936069 23731509 19189423 28552780 22580951 30157377 23917264 26217707 29764946 26163025 24929230 21478890 27354520 8754679 10048488 27862469 14593172 8852856 11449050 19245862 18507865 16188032 16381971 23299411
EMBL
BABH01015634
BABH01015635
JTDY01002438
KOB71424.1
NWSH01000334
PCG77316.1
+ More
KZ149922 PZC77714.1 ODYU01005843 SOQ47132.1 AGBW02013269 OWR43718.1 KB454495 EME31001.1 AC087192 DP000086 AP008216 AP014966 CM000147 AK070531 X15864 AWUE01014588 OMP02754.1 AWWV01007708 OMO95177.1 CM008054 PAN46522.1 CM009757 PUZ38318.1 PQIB02000001 RLN41725.1 PQIB02000005 RLN16698.1 EU952376 CM000781 ACG24494.1 AQK64663.1 KK198756 KCW78362.1 KD511611 EMT27233.1 AK354695 BAJ85914.1 KD062672 EMS63944.1 KC775782 AHE76167.1 CM000882 KQJ97425.1 CM001743 KJB24092.1 KZ647427 PPD91531.1 KN417221 KHG20985.1 LR031880 VDD60990.1 BT061870 CM007647 ACN26567.1 ONM05198.1 CM001881 EOY23776.1 KL596623 KER33689.1 UZAI01000358 VDO51757.1 CM000760 EER93954.1 LWDX02059504 OEL17580.1 KM510381 AIZ68150.1 JN204912 AEM89276.1 MVGT01004545 OUZ99073.1 CM008050 PAN26390.1 KX268094 ASF57549.1 CM009753 PUZ52873.1 LWDX02039652 OEL24463.1 PQIB02000016 PQIB02000004 RLM61306.1 RLN24513.1 AGNK02003468 CM003532 RCV29187.1 GU290545 GU290546 ADB80031.1 MVGT01004453 OUZ99434.1 MH450176 AXY40445.1 UZAK01033022 VDP33680.1 JXTC01000061 PON93040.1 KV894229 KER33688.1 OON18392.1 OIVN01000069 SPC73691.1 KM510380 AIZ68149.1 BT039716 EU966244 EU969279 CM000780 ACF84721.1 ACG38362.1 AQK58879.1 GBRH01216154 JAD81741.1 CM008048 PAN19295.1 CM009751 PUZ65790.1 PQIB02000009 PQIB02000003 RLN01079.1 RLN30125.1 MBFR01000003 PVU98105.1 CM000764 EES08028.1 LC040883 BAR40316.1 AB894322 BAO20877.1 JXTB01000434 PON40670.1 MBFS01002648 PVU94338.1 GU181353 ADQ27230.1 GU181354 ADQ27231.1 EU009452 ABV59397.1 HQ853237 ADW83713.1 CM000782 AQK79525.1 CM000768 EES19603.1 GL348718 EFH47645.1 LUHQ01000005 OAO92348.1 U37281 U27811 AB016893 CP002688 AY062702 AY063980 AY096397 AY114679 AY120779 JN983213 AFC88818.1 AY550991 EU284857 AAT45848.1 ACF06478.1 KU720631 AOF40434.1 MTKT01004810 PGOL01000328 OWM70058.1 PKI72382.1 AY742219 AAU93346.1 EES18852.1 KXG21029.1 BT033677 BT039238 EU961034 EU969521 BT054543 BT064389 BT066790 CM000784 ACF78682.1 ACG33152.1 AQK79519.1 AQK79526.1 AQK95624.1 AQK95628.1 CM000136 EEC67736.1 AY212324 AC120533 DP000010 AP008217 AK060893 AP014967 CM000148 AAO62546.1 AAX95098.1 ABA91666.1 BAF27675.1 BAG87600.1 BAT12814.1 EEE51698.1
KZ149922 PZC77714.1 ODYU01005843 SOQ47132.1 AGBW02013269 OWR43718.1 KB454495 EME31001.1 AC087192 DP000086 AP008216 AP014966 CM000147 AK070531 X15864 AWUE01014588 OMP02754.1 AWWV01007708 OMO95177.1 CM008054 PAN46522.1 CM009757 PUZ38318.1 PQIB02000001 RLN41725.1 PQIB02000005 RLN16698.1 EU952376 CM000781 ACG24494.1 AQK64663.1 KK198756 KCW78362.1 KD511611 EMT27233.1 AK354695 BAJ85914.1 KD062672 EMS63944.1 KC775782 AHE76167.1 CM000882 KQJ97425.1 CM001743 KJB24092.1 KZ647427 PPD91531.1 KN417221 KHG20985.1 LR031880 VDD60990.1 BT061870 CM007647 ACN26567.1 ONM05198.1 CM001881 EOY23776.1 KL596623 KER33689.1 UZAI01000358 VDO51757.1 CM000760 EER93954.1 LWDX02059504 OEL17580.1 KM510381 AIZ68150.1 JN204912 AEM89276.1 MVGT01004545 OUZ99073.1 CM008050 PAN26390.1 KX268094 ASF57549.1 CM009753 PUZ52873.1 LWDX02039652 OEL24463.1 PQIB02000016 PQIB02000004 RLM61306.1 RLN24513.1 AGNK02003468 CM003532 RCV29187.1 GU290545 GU290546 ADB80031.1 MVGT01004453 OUZ99434.1 MH450176 AXY40445.1 UZAK01033022 VDP33680.1 JXTC01000061 PON93040.1 KV894229 KER33688.1 OON18392.1 OIVN01000069 SPC73691.1 KM510380 AIZ68149.1 BT039716 EU966244 EU969279 CM000780 ACF84721.1 ACG38362.1 AQK58879.1 GBRH01216154 JAD81741.1 CM008048 PAN19295.1 CM009751 PUZ65790.1 PQIB02000009 PQIB02000003 RLN01079.1 RLN30125.1 MBFR01000003 PVU98105.1 CM000764 EES08028.1 LC040883 BAR40316.1 AB894322 BAO20877.1 JXTB01000434 PON40670.1 MBFS01002648 PVU94338.1 GU181353 ADQ27230.1 GU181354 ADQ27231.1 EU009452 ABV59397.1 HQ853237 ADW83713.1 CM000782 AQK79525.1 CM000768 EES19603.1 GL348718 EFH47645.1 LUHQ01000005 OAO92348.1 U37281 U27811 AB016893 CP002688 AY062702 AY063980 AY096397 AY114679 AY120779 JN983213 AFC88818.1 AY550991 EU284857 AAT45848.1 ACF06478.1 KU720631 AOF40434.1 MTKT01004810 PGOL01000328 OWM70058.1 PKI72382.1 AY742219 AAU93346.1 EES18852.1 KXG21029.1 BT033677 BT039238 EU961034 EU969521 BT054543 BT064389 BT066790 CM000784 ACF78682.1 ACG33152.1 AQK79519.1 AQK79526.1 AQK95624.1 AQK95628.1 CM000136 EEC67736.1 AY212324 AC120533 DP000010 AP008217 AK060893 AP014967 CM000148 AAO62546.1 AAX95098.1 ABA91666.1 BAF27675.1 BAG87600.1 BAT12814.1 EEE51698.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000030680
UP000032180
+ More
UP000026960 UP000006591 UP000008021 UP000008022 UP000026961 UP000026962 UP000059680 UP000187203 UP000188268 UP000243499 UP000244336 UP000006038 UP000275267 UP000007305 UP000030711 UP000019116 UP000011116 UP000015106 UP000008810 UP000032304 UP000239050 UP000189702 UP000012960 UP000032141 UP000026915 UP000050790 UP000277204 UP000000768 UP000195402 UP000004995 UP000050789 UP000237000 UP000228380 UP000245383 UP000237105 UP000245609 UP000008694 UP000078284 UP000006548 UP000233551 UP000007015
UP000026960 UP000006591 UP000008021 UP000008022 UP000026961 UP000026962 UP000059680 UP000187203 UP000188268 UP000243499 UP000244336 UP000006038 UP000275267 UP000007305 UP000030711 UP000019116 UP000011116 UP000015106 UP000008810 UP000032304 UP000239050 UP000189702 UP000012960 UP000032141 UP000026915 UP000050790 UP000277204 UP000000768 UP000195402 UP000004995 UP000050789 UP000237000 UP000228380 UP000245383 UP000237105 UP000245609 UP000008694 UP000078284 UP000006548 UP000233551 UP000007015
Pfam
PF00022 Actin
ProteinModelPortal
H9IXU9
A0A0L7L7V8
A0A2A4JZW6
A0A2W1BVQ8
A0A2H1W2S8
A0A212EQF7
+ More
M2W5E2 A0A0D9XLE8 A0A0D3HFI5 A0A0E0H4X6 A0A0E0EZS7 A0A0E0R185 A0A0E0BCK4 A0A0E0M9L0 A3C6D7 P0C539 A0A1R3K6S2 A0A1R3JK78 A0A2S3IKW4 A0A2T7C4P4 J3N402 A0A3L6TQH0 A0A3L6S6Y4 B6SI11 A0A059CIZ9 W5AH12 M8BZM2 F2CSU3 M8ASF1 A0A067YNJ5 I1I5C8 A0A0D2MXJ8 A0A2P5RXK1 A0A0B0P7F0 A0A1U8NDN5 M0TX26 A0A0D3CS69 A0A3P6G7D2 C0HHR4 A0A061G2I3 A0A075AJN4 A0A183LCR5 C5WW94 A0A1E5UXJ2 A0A0A7LUI4 G3KF67 A0A200PLG9 A0A2S3HMU2 A0A2U7N5E0 A0A2T7DBC6 A0A1E5VH52 A0A3L6PSP8 K3XQU0 D3JTC9 A0A200PMK0 A0A1U8JZ36 A0A385HDR9 A0A183K206 A0A2P5F5I2 A0A075A1W9 A0A2N9EGA0 A0A0A7LWK3 B4FRH8 A0A2H3Z634 A0A0A9D1Q4 A0A2S3HBM7 A0A2T7ED87 A0A3L6RE42 A0A2T9Z0G9 C5Y567 A0A0E3VJX4 V5YU14 M0TQW8 M0SBB4 A0A2P5AVT0 A0A2T9YPQ8 K7N8I1 K7N907 B4XAN5 E9P1J6 A0A1D6LI44 C5YYR1 D7M276 A0A178UEW4 P53492 H9B8D9 A0A2H3XM37 Q5J1K2 A0A1B3IJS2 A0A218WC81 Q5XPX5 C5YY31 B4F989 A0A0D3HIK8 B8BJ82 A0A0E0MCT4 J3N6A3 A0A0E0F2V9 A0A0D9XPE5 Q67G20
M2W5E2 A0A0D9XLE8 A0A0D3HFI5 A0A0E0H4X6 A0A0E0EZS7 A0A0E0R185 A0A0E0BCK4 A0A0E0M9L0 A3C6D7 P0C539 A0A1R3K6S2 A0A1R3JK78 A0A2S3IKW4 A0A2T7C4P4 J3N402 A0A3L6TQH0 A0A3L6S6Y4 B6SI11 A0A059CIZ9 W5AH12 M8BZM2 F2CSU3 M8ASF1 A0A067YNJ5 I1I5C8 A0A0D2MXJ8 A0A2P5RXK1 A0A0B0P7F0 A0A1U8NDN5 M0TX26 A0A0D3CS69 A0A3P6G7D2 C0HHR4 A0A061G2I3 A0A075AJN4 A0A183LCR5 C5WW94 A0A1E5UXJ2 A0A0A7LUI4 G3KF67 A0A200PLG9 A0A2S3HMU2 A0A2U7N5E0 A0A2T7DBC6 A0A1E5VH52 A0A3L6PSP8 K3XQU0 D3JTC9 A0A200PMK0 A0A1U8JZ36 A0A385HDR9 A0A183K206 A0A2P5F5I2 A0A075A1W9 A0A2N9EGA0 A0A0A7LWK3 B4FRH8 A0A2H3Z634 A0A0A9D1Q4 A0A2S3HBM7 A0A2T7ED87 A0A3L6RE42 A0A2T9Z0G9 C5Y567 A0A0E3VJX4 V5YU14 M0TQW8 M0SBB4 A0A2P5AVT0 A0A2T9YPQ8 K7N8I1 K7N907 B4XAN5 E9P1J6 A0A1D6LI44 C5YYR1 D7M276 A0A178UEW4 P53492 H9B8D9 A0A2H3XM37 Q5J1K2 A0A1B3IJS2 A0A218WC81 Q5XPX5 C5YY31 B4F989 A0A0D3HIK8 B8BJ82 A0A0E0MCT4 J3N6A3 A0A0E0F2V9 A0A0D9XPE5 Q67G20
PDB
4EFH
E-value=3.89152e-89,
Score=836
Ontologies
GO
PANTHER
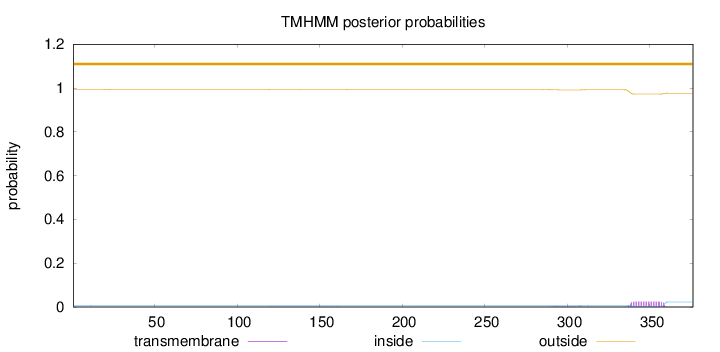
Topology
Subcellular location
Length:
376
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.5686
Exp number, first 60 AAs:
0.00114
Total prob of N-in:
0.00723
outside
1 - 376
Population Genetic Test Statistics
Pi
19.020718
Theta
23.048772
Tajima's D
-0.907205
CLR
0.087464
CSRT
0.158842057897105
Interpretation
Uncertain
