Gene
KWMTBOMO00027
Pre Gene Modal
BGIBMGA002051
Annotation
PREDICTED:_dipeptidase_1-like_[Bombyx_mori]
Full name
Dipeptidase
Location in the cell
Cytoplasmic Reliability : 2.232
Sequence
CDS
ATGGCTCCAACGGAGCGTGTTTTCTGGTCGGCGTTCGTGCCGTGCGGTACCCAAAACAGAGACGCCGTCCAGCTGACTCTGGAGCAGATTGACGTCATCAAGCGACTTGTCGAAAAATACCCTCAGCAGTTGCAATTCGCTACCTCCGTAACTGATATCTTGCAGTCGCACGGTGCTCGCCCACGTAAGATAGCATCCCTGATTGGCATCGAAGGAGGTCACTCTATCGCGAACTCTCTAGCAGTCCTCCGGAGCTACTACCAGCTTGGGGTTCGGTATATGACCCTCACTCACACCTGCCACACACCCTGGGCGGACTCCGCGAACGAGGCTCCAGCAGCGAACGGACTGACGGACTTCGGAGAGAAAGTGGTCCGCGAGATGAACCGCCTCGGTATGATCGTTGATCTGTCTCACGTGGGGGAGAACTCAACCAGAGCTGCGATCAAGCTGTCGAAGGCCCCCGTCATCTTCAGTCACTCCTCTGTCTACAGCCTCTGCAACCATAACCGAAACGTACCGGATGATATTATAAAATCATTGAAAGACAACGGAGGCATCATAATGGTGAACTTCTTCCCAGACTTCGTGAAGTGTGCTCCTAACGCCACACTTTCCGATGTTGCAGAGCATTTCCACCACATCAAGAGATTGGTTGGTGCTGACTACGTAGGTATCGGCGGAGACTTCGACGGCGTCAACAGGGTTCCCCGTGGCTTGGAAGATGTGTCAAAATACCCAGAACTGTTTGCTGAACTGCTTAGGAGTGGACAGTGGAGCGTTCAGGAGTTAAAGAACTTGGCCGGCCTCAATATGCTACGTGTGATGCGACAGGTGGAAAAGGTCCGCGACGAAATGCGAACAAACGGTGTCGAGCCTGAGGAGCACCCGGACTCGCCCAGCGACAACGGCAACTGCACCAGCAATGCTTTTTACACAGAGAACGTTTAA
Protein
MAPTERVFWSAFVPCGTQNRDAVQLTLEQIDVIKRLVEKYPQQLQFATSVTDILQSHGARPRKIASLIGIEGGHSIANSLAVLRSYYQLGVRYMTLTHTCHTPWADSANEAPAANGLTDFGEKVVREMNRLGMIVDLSHVGENSTRAAIKLSKAPVIFSHSSVYSLCNHNRNVPDDIIKSLKDNGGIIMVNFFPDFVKCAPNATLSDVAEHFHHIKRLVGADYVGIGGDFDGVNRVPRGLEDVSKYPELFAELLRSGQWSVQELKNLAGLNMLRVMRQVEKVRDEMRTNGVEPEEHPDSPSDNGNCTSNAFYTENV
Summary
Catalytic Activity
Hydrolysis of dipeptides.
Cofactor
Zn(2+)
Subunit
Homodimer; disulfide-linked.
Similarity
Belongs to the metallo-dependent hydrolases superfamily. Peptidase M19 family.
Feature
chain Dipeptidase
Uniprot
A0A194QXR7
A0A194PLX8
A0A212EQH0
A0A2A4K054
A0A195DKD9
A0A195D7C9
+ More
E9IBZ0 A0A195FVI0 A0A195BHJ7 A0A158P190 E2BQT2 A0A088A5Q6 A0A2A3E3F6 A0A0L7R9C5 K7IUE4 E0VEU5 A0A2J7R9X5 A0A3L8E3R7 B4Q8F2 A0A026WFU2 T1HCB5 U4UAM4 A0A0Q9WBB6 A0A3B0JZC6 A0A0Q9X6N7 A0A0R3NSJ4 Q179X0 A0A0P8Y317 A0A0B8RR20 Q9VJ94 A0A0J9R433 X2JA84 A0A0Q5VL41 A0A0J9TQF9 A0A182RQH4 B3NMC5 A0A098M0P0 A0A182LYM0 A0A182JMV9 A0A182JTK7 A0A182P3S6 A0A182Y6M4 A0A182VM54 A0A182XAL8 Q7QCH9 A0A182HSF9 A0A182W0S2 A0A3B3UQG4 A0A139WH56 A0A3B3Z3Y1 A0A182NRD1 A0A182QJ27 Q08BQ0 A0A087XHZ5 A0A0R1DV33 F1QFU9 A0A182FPA6 A0A336M0A2 V8P4B3 A0A1W4VHE8 B4NKQ8 A0A0P5V9G1 I3J4I1 A0A067QRU6 A0A1I8Q540 A0A3P8R8N7 W5KI72 A0A1J1IBU3 A0A1W4ZWP7 A0A087ZTV9 A0A3P9QGR9 A0A3P9AY36 W8AEG4 E0VS36 A0A3L8D6Q7 A0A2A3EG90 A0A3P9AC48 A0A2I4ASR4 A0A0K8RVV8 B4PSV0 Q297B0 B4GED4 A0A3B4DVB7 H9JQI3 A0A3B0JLU8 A0A3B4C3A3 A0A2J7RAA8 W5J7N1 A0A067RD78 A0A0P4W9D6 A0A2H1X0W8 A0A0L0CAK9 A0A2J7QYI1 A7SK00 A0A1W4VML0 Q9VBG1 B3P685 A0A1S3PTE2 A0A0L7RES1
E9IBZ0 A0A195FVI0 A0A195BHJ7 A0A158P190 E2BQT2 A0A088A5Q6 A0A2A3E3F6 A0A0L7R9C5 K7IUE4 E0VEU5 A0A2J7R9X5 A0A3L8E3R7 B4Q8F2 A0A026WFU2 T1HCB5 U4UAM4 A0A0Q9WBB6 A0A3B0JZC6 A0A0Q9X6N7 A0A0R3NSJ4 Q179X0 A0A0P8Y317 A0A0B8RR20 Q9VJ94 A0A0J9R433 X2JA84 A0A0Q5VL41 A0A0J9TQF9 A0A182RQH4 B3NMC5 A0A098M0P0 A0A182LYM0 A0A182JMV9 A0A182JTK7 A0A182P3S6 A0A182Y6M4 A0A182VM54 A0A182XAL8 Q7QCH9 A0A182HSF9 A0A182W0S2 A0A3B3UQG4 A0A139WH56 A0A3B3Z3Y1 A0A182NRD1 A0A182QJ27 Q08BQ0 A0A087XHZ5 A0A0R1DV33 F1QFU9 A0A182FPA6 A0A336M0A2 V8P4B3 A0A1W4VHE8 B4NKQ8 A0A0P5V9G1 I3J4I1 A0A067QRU6 A0A1I8Q540 A0A3P8R8N7 W5KI72 A0A1J1IBU3 A0A1W4ZWP7 A0A087ZTV9 A0A3P9QGR9 A0A3P9AY36 W8AEG4 E0VS36 A0A3L8D6Q7 A0A2A3EG90 A0A3P9AC48 A0A2I4ASR4 A0A0K8RVV8 B4PSV0 Q297B0 B4GED4 A0A3B4DVB7 H9JQI3 A0A3B0JLU8 A0A3B4C3A3 A0A2J7RAA8 W5J7N1 A0A067RD78 A0A0P4W9D6 A0A2H1X0W8 A0A0L0CAK9 A0A2J7QYI1 A7SK00 A0A1W4VML0 Q9VBG1 B3P685 A0A1S3PTE2 A0A0L7RES1
EC Number
3.4.13.19
Pubmed
26354079
22118469
21282665
21347285
20798317
20075255
+ More
20566863 30249741 17994087 24508170 23537049 18057021 15632085 23185243 17510324 25476704 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 25244985 12364791 14747013 17210077 18362917 19820115 17550304 23594743 24297900 25186727 24845553 25329095 24495485 25069045 25727380 19121390 20920257 23761445 26108605 17615350 26109357 26109356
20566863 30249741 17994087 24508170 23537049 18057021 15632085 23185243 17510324 25476704 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 25244985 12364791 14747013 17210077 18362917 19820115 17550304 23594743 24297900 25186727 24845553 25329095 24495485 25069045 25727380 19121390 20920257 23761445 26108605 17615350 26109357 26109356
EMBL
KQ460953
KPJ10333.1
KQ459599
KPI94332.1
AGBW02013269
OWR43719.1
+ More
NWSH01000334 PCG77308.1 KQ980765 KYN13353.1 KQ976749 KYN08762.1 GL762140 EFZ21903.1 KQ981272 KYN43869.1 KQ976488 KYM83641.1 ADTU01006019 ADTU01006020 ADTU01006021 GL449799 EFN81957.1 KZ288400 PBC26257.1 KQ414627 KOC67480.1 AAZX01005844 DS235098 EEB11901.1 NEVH01006580 PNF37626.1 QOIP01000001 RLU26869.1 CM000361 EDX05362.1 KK107242 EZA54536.1 ACPB03008996 KB631899 ERL87010.1 CH940649 KRF81945.1 OUUW01000004 SPP79029.1 CH933807 KRG03677.1 CH379060 KRT04136.1 CH477344 EAT43028.1 CH902620 KPU73057.1 GBSH01000861 JAG68164.1 AE014134 AAF53659.5 CM002910 KMY90786.1 AHN54546.1 CH954179 KQS61940.1 KMY90785.1 EDV54796.2 GBSI01000721 JAC95775.1 AXCM01000160 AAAB01008859 EAA08208.5 APCN01000271 KQ971343 KYB27246.1 AXCN02001466 BC124623 AAI24624.1 AYCK01005495 AYCK01005496 CM000158 KRJ99088.1 CR382281 CU468003 UFQT01000071 SSX19398.1 AZIM01000835 ETE69165.1 CH964272 EDW84119.2 GDIP01103280 JAM00435.1 AERX01013846 AERX01013847 KK853032 KDR12256.1 CVRI01000047 CRK97751.1 GAMC01019979 GAMC01019978 GAMC01019975 GAMC01019971 JAB86580.1 DS235742 EEB16192.1 QOIP01000012 RLU15871.1 KZ288254 PBC30728.1 GBKC01000994 JAG45076.1 CM000160 EDW98637.1 CM000070 EAL28296.2 CH479182 EDW33969.1 BABH01012020 OUUW01000007 SPP83284.1 NEVH01006576 PNF37768.1 ADMH02002018 ETN59976.1 KK852779 KDR16715.1 GDRN01070938 JAI63790.1 ODYU01012538 SOQ58912.1 JRES01000664 KNC29438.1 NEVH01009089 PNF33643.1 DS469682 EDO35966.1 AE014297 BT010258 BT031191 AAF56581.2 AAQ23576.1 ABY20432.1 CH954182 EDV53555.1 KQ414610 KOC69236.1
NWSH01000334 PCG77308.1 KQ980765 KYN13353.1 KQ976749 KYN08762.1 GL762140 EFZ21903.1 KQ981272 KYN43869.1 KQ976488 KYM83641.1 ADTU01006019 ADTU01006020 ADTU01006021 GL449799 EFN81957.1 KZ288400 PBC26257.1 KQ414627 KOC67480.1 AAZX01005844 DS235098 EEB11901.1 NEVH01006580 PNF37626.1 QOIP01000001 RLU26869.1 CM000361 EDX05362.1 KK107242 EZA54536.1 ACPB03008996 KB631899 ERL87010.1 CH940649 KRF81945.1 OUUW01000004 SPP79029.1 CH933807 KRG03677.1 CH379060 KRT04136.1 CH477344 EAT43028.1 CH902620 KPU73057.1 GBSH01000861 JAG68164.1 AE014134 AAF53659.5 CM002910 KMY90786.1 AHN54546.1 CH954179 KQS61940.1 KMY90785.1 EDV54796.2 GBSI01000721 JAC95775.1 AXCM01000160 AAAB01008859 EAA08208.5 APCN01000271 KQ971343 KYB27246.1 AXCN02001466 BC124623 AAI24624.1 AYCK01005495 AYCK01005496 CM000158 KRJ99088.1 CR382281 CU468003 UFQT01000071 SSX19398.1 AZIM01000835 ETE69165.1 CH964272 EDW84119.2 GDIP01103280 JAM00435.1 AERX01013846 AERX01013847 KK853032 KDR12256.1 CVRI01000047 CRK97751.1 GAMC01019979 GAMC01019978 GAMC01019975 GAMC01019971 JAB86580.1 DS235742 EEB16192.1 QOIP01000012 RLU15871.1 KZ288254 PBC30728.1 GBKC01000994 JAG45076.1 CM000160 EDW98637.1 CM000070 EAL28296.2 CH479182 EDW33969.1 BABH01012020 OUUW01000007 SPP83284.1 NEVH01006576 PNF37768.1 ADMH02002018 ETN59976.1 KK852779 KDR16715.1 GDRN01070938 JAI63790.1 ODYU01012538 SOQ58912.1 JRES01000664 KNC29438.1 NEVH01009089 PNF33643.1 DS469682 EDO35966.1 AE014297 BT010258 BT031191 AAF56581.2 AAQ23576.1 ABY20432.1 CH954182 EDV53555.1 KQ414610 KOC69236.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000218220
UP000078492
UP000078542
+ More
UP000078541 UP000078540 UP000005205 UP000008237 UP000005203 UP000242457 UP000053825 UP000002358 UP000009046 UP000235965 UP000279307 UP000000304 UP000053097 UP000015103 UP000030742 UP000008792 UP000268350 UP000009192 UP000001819 UP000008820 UP000007801 UP000000803 UP000008711 UP000075900 UP000075883 UP000075880 UP000075881 UP000075885 UP000076408 UP000075903 UP000076407 UP000007062 UP000075840 UP000075920 UP000261500 UP000007266 UP000261480 UP000075884 UP000075886 UP000028760 UP000002282 UP000000437 UP000069272 UP000192221 UP000007798 UP000005207 UP000027135 UP000095300 UP000265100 UP000018467 UP000183832 UP000192224 UP000242638 UP000265160 UP000265140 UP000192220 UP000008744 UP000261440 UP000005204 UP000000673 UP000037069 UP000001593 UP000087266
UP000078541 UP000078540 UP000005205 UP000008237 UP000005203 UP000242457 UP000053825 UP000002358 UP000009046 UP000235965 UP000279307 UP000000304 UP000053097 UP000015103 UP000030742 UP000008792 UP000268350 UP000009192 UP000001819 UP000008820 UP000007801 UP000000803 UP000008711 UP000075900 UP000075883 UP000075880 UP000075881 UP000075885 UP000076408 UP000075903 UP000076407 UP000007062 UP000075840 UP000075920 UP000261500 UP000007266 UP000261480 UP000075884 UP000075886 UP000028760 UP000002282 UP000000437 UP000069272 UP000192221 UP000007798 UP000005207 UP000027135 UP000095300 UP000265100 UP000018467 UP000183832 UP000192224 UP000242638 UP000265160 UP000265140 UP000192220 UP000008744 UP000261440 UP000005204 UP000000673 UP000037069 UP000001593 UP000087266
Pfam
PF01244 Peptidase_M19
SUPFAM
SSF51556
SSF51556
CDD
ProteinModelPortal
A0A194QXR7
A0A194PLX8
A0A212EQH0
A0A2A4K054
A0A195DKD9
A0A195D7C9
+ More
E9IBZ0 A0A195FVI0 A0A195BHJ7 A0A158P190 E2BQT2 A0A088A5Q6 A0A2A3E3F6 A0A0L7R9C5 K7IUE4 E0VEU5 A0A2J7R9X5 A0A3L8E3R7 B4Q8F2 A0A026WFU2 T1HCB5 U4UAM4 A0A0Q9WBB6 A0A3B0JZC6 A0A0Q9X6N7 A0A0R3NSJ4 Q179X0 A0A0P8Y317 A0A0B8RR20 Q9VJ94 A0A0J9R433 X2JA84 A0A0Q5VL41 A0A0J9TQF9 A0A182RQH4 B3NMC5 A0A098M0P0 A0A182LYM0 A0A182JMV9 A0A182JTK7 A0A182P3S6 A0A182Y6M4 A0A182VM54 A0A182XAL8 Q7QCH9 A0A182HSF9 A0A182W0S2 A0A3B3UQG4 A0A139WH56 A0A3B3Z3Y1 A0A182NRD1 A0A182QJ27 Q08BQ0 A0A087XHZ5 A0A0R1DV33 F1QFU9 A0A182FPA6 A0A336M0A2 V8P4B3 A0A1W4VHE8 B4NKQ8 A0A0P5V9G1 I3J4I1 A0A067QRU6 A0A1I8Q540 A0A3P8R8N7 W5KI72 A0A1J1IBU3 A0A1W4ZWP7 A0A087ZTV9 A0A3P9QGR9 A0A3P9AY36 W8AEG4 E0VS36 A0A3L8D6Q7 A0A2A3EG90 A0A3P9AC48 A0A2I4ASR4 A0A0K8RVV8 B4PSV0 Q297B0 B4GED4 A0A3B4DVB7 H9JQI3 A0A3B0JLU8 A0A3B4C3A3 A0A2J7RAA8 W5J7N1 A0A067RD78 A0A0P4W9D6 A0A2H1X0W8 A0A0L0CAK9 A0A2J7QYI1 A7SK00 A0A1W4VML0 Q9VBG1 B3P685 A0A1S3PTE2 A0A0L7RES1
E9IBZ0 A0A195FVI0 A0A195BHJ7 A0A158P190 E2BQT2 A0A088A5Q6 A0A2A3E3F6 A0A0L7R9C5 K7IUE4 E0VEU5 A0A2J7R9X5 A0A3L8E3R7 B4Q8F2 A0A026WFU2 T1HCB5 U4UAM4 A0A0Q9WBB6 A0A3B0JZC6 A0A0Q9X6N7 A0A0R3NSJ4 Q179X0 A0A0P8Y317 A0A0B8RR20 Q9VJ94 A0A0J9R433 X2JA84 A0A0Q5VL41 A0A0J9TQF9 A0A182RQH4 B3NMC5 A0A098M0P0 A0A182LYM0 A0A182JMV9 A0A182JTK7 A0A182P3S6 A0A182Y6M4 A0A182VM54 A0A182XAL8 Q7QCH9 A0A182HSF9 A0A182W0S2 A0A3B3UQG4 A0A139WH56 A0A3B3Z3Y1 A0A182NRD1 A0A182QJ27 Q08BQ0 A0A087XHZ5 A0A0R1DV33 F1QFU9 A0A182FPA6 A0A336M0A2 V8P4B3 A0A1W4VHE8 B4NKQ8 A0A0P5V9G1 I3J4I1 A0A067QRU6 A0A1I8Q540 A0A3P8R8N7 W5KI72 A0A1J1IBU3 A0A1W4ZWP7 A0A087ZTV9 A0A3P9QGR9 A0A3P9AY36 W8AEG4 E0VS36 A0A3L8D6Q7 A0A2A3EG90 A0A3P9AC48 A0A2I4ASR4 A0A0K8RVV8 B4PSV0 Q297B0 B4GED4 A0A3B4DVB7 H9JQI3 A0A3B0JLU8 A0A3B4C3A3 A0A2J7RAA8 W5J7N1 A0A067RD78 A0A0P4W9D6 A0A2H1X0W8 A0A0L0CAK9 A0A2J7QYI1 A7SK00 A0A1W4VML0 Q9VBG1 B3P685 A0A1S3PTE2 A0A0L7RES1
PDB
1ITU
E-value=1.3977e-87,
Score=822
Ontologies
GO
PANTHER
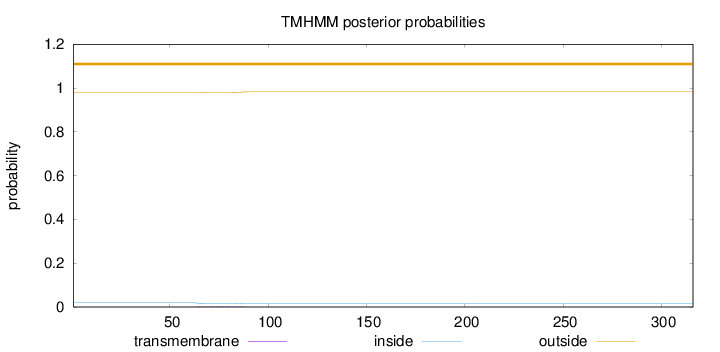
Topology
Subcellular location
Membrane
Length:
316
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0780399999999998
Exp number, first 60 AAs:
0.00036
Total prob of N-in:
0.01910
outside
1 - 316
Population Genetic Test Statistics
Pi
18.964123
Theta
23.110051
Tajima's D
-1.496208
CLR
0
CSRT
0.0599970001499925
Interpretation
Uncertain
