Gene
KWMTBOMO00026
Pre Gene Modal
BGIBMGA002081
Annotation
Mitochondrial_processing_peptidase_beta_subunit_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.095 Mitochondrial Reliability : 1.06
Sequence
CDS
ATGTTTCGTAAAACGTTCAAAAGTTTTTTTCCAAAATGGATAAGGTTAAATAGGAAACGCTCTGCTGGAGTCTGTCATCCTGTTGATCCAAATGATCCTGGTACCAAGATCAGTCTCCTACCGAATGGAGTGCGGATTGCAACTGAACAGACGCAATCACCACTTGCTTGCGTTTCTCTGTTCATCGAAGCAGGCCCGCGTTTTGAAACTCCGGAAAACAACGGTGCTAGCCATTTTCTAGAACACATGGCATTCTGCGGTTTTAAGTCAATGAACCAATGTGAAATAGAACATAATCTTTTGCAAATGGGCGCGAAGATAAACGCAGAAACAACTAAAGAGATACAAAGATTTGTGGCGATTTGTCCATCAGAAAATGCGCACGAGATGGTCGCTTTTCTTTGTCGCATTATAACTGACCTTGACCTAAATGACTCTTCAATAGAGACCGAAAAGCACAACATGTGTTACGAACTAGTGGACTCCGATAATGATCCTAAAGCTGTTATGTTCGAATATTTGCATCAAACTGCTTTTCAAGGAACCCCGCTGGCTCAAAGTGTCATAGGCCCATCGAAGAATATTCAAAACTTTGACAGTCAACTTCTGTCATCCTTTATGACCGATCATTACCAGCCTTATAAAGTTTGCTTTGCTACATCTGGAAACGTGGACCACAAAGAAGTCGTACGCATCGCAGAAACGATGTGCGGAAAAATGGTTGGAGATCCAACGAAGCAGTTTGGAAGAGGCCCTTGCCGATTTACTGGGTCCCAGATTATGTACCGTGACGATTCTATGCCATGCGCTCACGTTGCCATTGGGTTCGAAGTCCCGGGATATGGCCATGAAGATTACTTAAAGCTGTTGGTAATGGGGTGTATGATGGGAGCCTGGGATAAAAGTCAAGGAGGAGGTAACAGTAACGTACCAGTACTGGCCTGCGCTGCTGCCTCTGGTCTTTGCGAGAGCTACGAGCCGTTTTATTTTCCATACGGCGATATCGGCTTATGGGGTGTGTACTATGTGGGACAGCCACTAGTTTTAGAAGATATGTTAAACAACATTCAGGACTACTGGATGAAGATGTGCGTTAGTGTTCATTATACAGATTTAGAAAGGGCGAAAAACTTGGCCAAACTAAAGGTAGCGAAGATGTTTGATGGAACCGTGAATTCCTCGTATGATATTGGAATGCAGTTGATGTATTTTTGTGGAAGGAAACCGTTAATTAGTCAATATGAATTGTTGTCTAATATAAAGCCAGACTCGATCAGAGAAGCTGCTGATAAATATTTGTACGACAGGTGTCCGGCGGTAGCCTGTGTGGGGCCCACGGAGGGCATGCCCGACTACACGAAAATCAGAGCAGGACAATATTGGTTGAGATACTGA
Protein
MFRKTFKSFFPKWIRLNRKRSAGVCHPVDPNDPGTKISLLPNGVRIATEQTQSPLACVSLFIEAGPRFETPENNGASHFLEHMAFCGFKSMNQCEIEHNLLQMGAKINAETTKEIQRFVAICPSENAHEMVAFLCRIITDLDLNDSSIETEKHNMCYELVDSDNDPKAVMFEYLHQTAFQGTPLAQSVIGPSKNIQNFDSQLLSSFMTDHYQPYKVCFATSGNVDHKEVVRIAETMCGKMVGDPTKQFGRGPCRFTGSQIMYRDDSMPCAHVAIGFEVPGYGHEDYLKLLVMGCMMGAWDKSQGGGNSNVPVLACAAASGLCESYEPFYFPYGDIGLWGVYYVGQPLVLEDMLNNIQDYWMKMCVSVHYTDLERAKNLAKLKVAKMFDGTVNSSYDIGMQLMYFCGRKPLISQYELLSNIKPDSIREAADKYLYDRCPAVACVGPTEGMPDYTKIRAGQYWLRY
Summary
Similarity
Belongs to the peptidase M16 family.
Uniprot
H9IXU8
A0A0L7KUH5
A0A2H1W690
A0A194QZF2
A0A2A4K0W0
A0A212EQF1
+ More
A0A194PNP0 I4DK27 A0A194QVC2 A0A2H1VE55 A0A2A4JTD4 A0A0L7LMY8 A0A1L8DTM4 A0A182WQY9 A0A0K8TS24 D3TRB5 A0A2M4A748 W5JN85 A0A2M4A819 A0A224XJ82 A0A1I8NRV4 T1DMV8 A0A2M3Z0P8 A0A1A9WYN4 A0A161MLQ6 B4G6F2 A0A0L0CB92 A0A182FIQ4 D3TRB1 A0A2M4BLW9 A0A1A9ZZ69 Q29AI0 A0A1A9YT06 A0A1B0B3C0 A0A1L8EEG7 A0A182J6J8 A0A023F9T0 A0A3B0KHU6 A0A182VKS0 A0A182TZE6 A0A182X960 Q7PSV0 A0A182QFE4 A0A084VIU0 A0A0P4VUN1 R4G4U9 A0A1A9VFS3 A0A182T1J9 A0A182Y4W8 A0A182MA04 A0A0V0G3H4 B4JF35 A0A182IDK5 A0A0M4F4P0 T1PD60 B4PSN4 A0A1W4VMN1 B4QZ72 B4HE39 Q9VFF0 B3P3P8 A0A0A1XFT2 A0A240SWL7 A0A240SXE2 A0A182PL50 B4NJ55 B4M411 A0A182N0I7 U5ETG1 B4K945 A0A067R6C1 A0A182K790 B3LWQ6 A0A1B0FI28 A0A034VJX9 W8B9H4 A0A232EKT3 A0A1W4X8N1 A0A023ESS2 Q17A09 A0A2J7PY28 A0A0P5IWE6 A0A164RZD6 A0A0N8ARF2 A0A0P4ZM21 A0A0N8B320 A0A0P5LT03 A0A0P5FU69 A0A0P5AUN9 A0A0P5K6G8 A0A084VRA3 A0A0P5FZP2 A0A0P5LR24 A0A0P5MNI0 K7IPF7 A0A0C9PY15 E9GEU2 A0A182HLT6 A0A182TZA9 A0A154PGJ5
A0A194PNP0 I4DK27 A0A194QVC2 A0A2H1VE55 A0A2A4JTD4 A0A0L7LMY8 A0A1L8DTM4 A0A182WQY9 A0A0K8TS24 D3TRB5 A0A2M4A748 W5JN85 A0A2M4A819 A0A224XJ82 A0A1I8NRV4 T1DMV8 A0A2M3Z0P8 A0A1A9WYN4 A0A161MLQ6 B4G6F2 A0A0L0CB92 A0A182FIQ4 D3TRB1 A0A2M4BLW9 A0A1A9ZZ69 Q29AI0 A0A1A9YT06 A0A1B0B3C0 A0A1L8EEG7 A0A182J6J8 A0A023F9T0 A0A3B0KHU6 A0A182VKS0 A0A182TZE6 A0A182X960 Q7PSV0 A0A182QFE4 A0A084VIU0 A0A0P4VUN1 R4G4U9 A0A1A9VFS3 A0A182T1J9 A0A182Y4W8 A0A182MA04 A0A0V0G3H4 B4JF35 A0A182IDK5 A0A0M4F4P0 T1PD60 B4PSN4 A0A1W4VMN1 B4QZ72 B4HE39 Q9VFF0 B3P3P8 A0A0A1XFT2 A0A240SWL7 A0A240SXE2 A0A182PL50 B4NJ55 B4M411 A0A182N0I7 U5ETG1 B4K945 A0A067R6C1 A0A182K790 B3LWQ6 A0A1B0FI28 A0A034VJX9 W8B9H4 A0A232EKT3 A0A1W4X8N1 A0A023ESS2 Q17A09 A0A2J7PY28 A0A0P5IWE6 A0A164RZD6 A0A0N8ARF2 A0A0P4ZM21 A0A0N8B320 A0A0P5LT03 A0A0P5FU69 A0A0P5AUN9 A0A0P5K6G8 A0A084VRA3 A0A0P5FZP2 A0A0P5LR24 A0A0P5MNI0 K7IPF7 A0A0C9PY15 E9GEU2 A0A182HLT6 A0A182TZA9 A0A154PGJ5
Pubmed
19121390
26227816
26354079
22118469
22651552
26369729
+ More
20353571 20920257 23761445 17994087 26108605 15632085 25474469 12364791 14747013 17210077 24438588 27129103 25244985 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 24845553 25348373 24495485 28648823 24945155 26483478 17510324 20075255 21292972
20353571 20920257 23761445 17994087 26108605 15632085 25474469 12364791 14747013 17210077 24438588 27129103 25244985 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 24845553 25348373 24495485 28648823 24945155 26483478 17510324 20075255 21292972
EMBL
BABH01015632
JTDY01005490
KOB66947.1
ODYU01006579
SOQ48543.1
KQ460953
+ More
KPJ10355.1 NWSH01000334 PCG77310.1 AGBW02013269 OWR43720.1 KQ459599 KPI94359.1 AK401645 BAM18267.1 KQ461103 KPJ09417.1 ODYU01002058 SOQ39097.1 NWSH01000617 PCG75271.1 JTDY01000495 KOB76918.1 GFDF01004350 JAV09734.1 GDAI01000451 JAI17152.1 EZ423967 ADD20243.1 GGFK01003137 MBW36458.1 ADMH02001041 ETN64350.1 GGFK01003623 MBW36944.1 GFTR01006548 JAW09878.1 GAMD01003405 JAA98185.1 GGFM01001339 MBW22090.1 GEMB01002202 JAS00978.1 CH479179 EDW23910.1 JRES01000641 KNC29683.1 EZ423963 ADD20239.1 GGFJ01004938 MBW54079.1 CM000070 EAL27370.1 JXJN01007808 GFDG01001711 JAV17088.1 GBBI01000953 JAC17759.1 OUUW01000013 SPP88030.1 AAAB01008811 EAA04978.5 AXCN02002488 ATLV01013404 KE524855 KFB37884.1 GDKW01000857 JAI55738.1 ACPB03023307 GAHY01000441 JAA77069.1 AXCM01001160 GECL01003480 JAP02644.1 CH916369 EDV93316.1 APCN01004712 CP012526 ALC46345.1 KA646050 AFP60679.1 CM000160 EDW97530.1 CM000364 EDX12900.1 CH480815 EDW42131.1 AE014297 AY058243 AAF55110.2 AAL13472.1 AAN13622.1 CH954181 EDV48936.1 GBXI01004480 JAD09812.1 JXJN01002847 CH964272 EDW83848.1 CH940652 EDW59372.1 GANO01002793 JAB57078.1 CH933806 EDW14458.1 KK852856 KDR14885.1 CH902617 EDV42694.1 CCAG010001331 GAKP01016525 JAC42427.1 GAMC01008640 JAB97915.1 NNAY01003732 OXU18931.1 JXUM01121409 GAPW01001258 KQ566497 JAC12340.1 KXJ70078.1 CH477341 EAT43098.1 EJY57583.1 NEVH01020851 PNF21226.1 GDIQ01232802 JAK18923.1 LRGB01002121 KZS09086.1 GDIQ01250243 JAK01482.1 GDIP01212779 JAJ10623.1 GDIQ01217699 JAK34026.1 GDIQ01166182 JAK85543.1 GDIQ01250244 JAK01481.1 GDIP01197742 JAJ25660.1 GDIQ01189962 JAK61763.1 ATLV01015566 KE525023 KFB40497.1 GDIQ01251689 JAK00036.1 GDIQ01176666 JAK75059.1 GDIQ01176665 JAK75060.1 GBYB01006418 JAG76185.1 GL732541 EFX81896.1 APCN01000915 KQ434899 KZC10985.1
KPJ10355.1 NWSH01000334 PCG77310.1 AGBW02013269 OWR43720.1 KQ459599 KPI94359.1 AK401645 BAM18267.1 KQ461103 KPJ09417.1 ODYU01002058 SOQ39097.1 NWSH01000617 PCG75271.1 JTDY01000495 KOB76918.1 GFDF01004350 JAV09734.1 GDAI01000451 JAI17152.1 EZ423967 ADD20243.1 GGFK01003137 MBW36458.1 ADMH02001041 ETN64350.1 GGFK01003623 MBW36944.1 GFTR01006548 JAW09878.1 GAMD01003405 JAA98185.1 GGFM01001339 MBW22090.1 GEMB01002202 JAS00978.1 CH479179 EDW23910.1 JRES01000641 KNC29683.1 EZ423963 ADD20239.1 GGFJ01004938 MBW54079.1 CM000070 EAL27370.1 JXJN01007808 GFDG01001711 JAV17088.1 GBBI01000953 JAC17759.1 OUUW01000013 SPP88030.1 AAAB01008811 EAA04978.5 AXCN02002488 ATLV01013404 KE524855 KFB37884.1 GDKW01000857 JAI55738.1 ACPB03023307 GAHY01000441 JAA77069.1 AXCM01001160 GECL01003480 JAP02644.1 CH916369 EDV93316.1 APCN01004712 CP012526 ALC46345.1 KA646050 AFP60679.1 CM000160 EDW97530.1 CM000364 EDX12900.1 CH480815 EDW42131.1 AE014297 AY058243 AAF55110.2 AAL13472.1 AAN13622.1 CH954181 EDV48936.1 GBXI01004480 JAD09812.1 JXJN01002847 CH964272 EDW83848.1 CH940652 EDW59372.1 GANO01002793 JAB57078.1 CH933806 EDW14458.1 KK852856 KDR14885.1 CH902617 EDV42694.1 CCAG010001331 GAKP01016525 JAC42427.1 GAMC01008640 JAB97915.1 NNAY01003732 OXU18931.1 JXUM01121409 GAPW01001258 KQ566497 JAC12340.1 KXJ70078.1 CH477341 EAT43098.1 EJY57583.1 NEVH01020851 PNF21226.1 GDIQ01232802 JAK18923.1 LRGB01002121 KZS09086.1 GDIQ01250243 JAK01482.1 GDIP01212779 JAJ10623.1 GDIQ01217699 JAK34026.1 GDIQ01166182 JAK85543.1 GDIQ01250244 JAK01481.1 GDIP01197742 JAJ25660.1 GDIQ01189962 JAK61763.1 ATLV01015566 KE525023 KFB40497.1 GDIQ01251689 JAK00036.1 GDIQ01176666 JAK75059.1 GDIQ01176665 JAK75060.1 GBYB01006418 JAG76185.1 GL732541 EFX81896.1 APCN01000915 KQ434899 KZC10985.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000218220
UP000007151
UP000053268
+ More
UP000075920 UP000000673 UP000095300 UP000091820 UP000008744 UP000037069 UP000069272 UP000092445 UP000001819 UP000092443 UP000092460 UP000075880 UP000268350 UP000075903 UP000075902 UP000076407 UP000007062 UP000075886 UP000030765 UP000015103 UP000078200 UP000075901 UP000076408 UP000075883 UP000001070 UP000075840 UP000092553 UP000095301 UP000002282 UP000192221 UP000000304 UP000001292 UP000000803 UP000008711 UP000075885 UP000007798 UP000008792 UP000075884 UP000009192 UP000027135 UP000075881 UP000007801 UP000092444 UP000215335 UP000192223 UP000069940 UP000249989 UP000008820 UP000235965 UP000076858 UP000002358 UP000000305 UP000076502
UP000075920 UP000000673 UP000095300 UP000091820 UP000008744 UP000037069 UP000069272 UP000092445 UP000001819 UP000092443 UP000092460 UP000075880 UP000268350 UP000075903 UP000075902 UP000076407 UP000007062 UP000075886 UP000030765 UP000015103 UP000078200 UP000075901 UP000076408 UP000075883 UP000001070 UP000075840 UP000092553 UP000095301 UP000002282 UP000192221 UP000000304 UP000001292 UP000000803 UP000008711 UP000075885 UP000007798 UP000008792 UP000075884 UP000009192 UP000027135 UP000075881 UP000007801 UP000092444 UP000215335 UP000192223 UP000069940 UP000249989 UP000008820 UP000235965 UP000076858 UP000002358 UP000000305 UP000076502
PRIDE
Interpro
SUPFAM
SSF63411
SSF63411
ProteinModelPortal
H9IXU8
A0A0L7KUH5
A0A2H1W690
A0A194QZF2
A0A2A4K0W0
A0A212EQF1
+ More
A0A194PNP0 I4DK27 A0A194QVC2 A0A2H1VE55 A0A2A4JTD4 A0A0L7LMY8 A0A1L8DTM4 A0A182WQY9 A0A0K8TS24 D3TRB5 A0A2M4A748 W5JN85 A0A2M4A819 A0A224XJ82 A0A1I8NRV4 T1DMV8 A0A2M3Z0P8 A0A1A9WYN4 A0A161MLQ6 B4G6F2 A0A0L0CB92 A0A182FIQ4 D3TRB1 A0A2M4BLW9 A0A1A9ZZ69 Q29AI0 A0A1A9YT06 A0A1B0B3C0 A0A1L8EEG7 A0A182J6J8 A0A023F9T0 A0A3B0KHU6 A0A182VKS0 A0A182TZE6 A0A182X960 Q7PSV0 A0A182QFE4 A0A084VIU0 A0A0P4VUN1 R4G4U9 A0A1A9VFS3 A0A182T1J9 A0A182Y4W8 A0A182MA04 A0A0V0G3H4 B4JF35 A0A182IDK5 A0A0M4F4P0 T1PD60 B4PSN4 A0A1W4VMN1 B4QZ72 B4HE39 Q9VFF0 B3P3P8 A0A0A1XFT2 A0A240SWL7 A0A240SXE2 A0A182PL50 B4NJ55 B4M411 A0A182N0I7 U5ETG1 B4K945 A0A067R6C1 A0A182K790 B3LWQ6 A0A1B0FI28 A0A034VJX9 W8B9H4 A0A232EKT3 A0A1W4X8N1 A0A023ESS2 Q17A09 A0A2J7PY28 A0A0P5IWE6 A0A164RZD6 A0A0N8ARF2 A0A0P4ZM21 A0A0N8B320 A0A0P5LT03 A0A0P5FU69 A0A0P5AUN9 A0A0P5K6G8 A0A084VRA3 A0A0P5FZP2 A0A0P5LR24 A0A0P5MNI0 K7IPF7 A0A0C9PY15 E9GEU2 A0A182HLT6 A0A182TZA9 A0A154PGJ5
A0A194PNP0 I4DK27 A0A194QVC2 A0A2H1VE55 A0A2A4JTD4 A0A0L7LMY8 A0A1L8DTM4 A0A182WQY9 A0A0K8TS24 D3TRB5 A0A2M4A748 W5JN85 A0A2M4A819 A0A224XJ82 A0A1I8NRV4 T1DMV8 A0A2M3Z0P8 A0A1A9WYN4 A0A161MLQ6 B4G6F2 A0A0L0CB92 A0A182FIQ4 D3TRB1 A0A2M4BLW9 A0A1A9ZZ69 Q29AI0 A0A1A9YT06 A0A1B0B3C0 A0A1L8EEG7 A0A182J6J8 A0A023F9T0 A0A3B0KHU6 A0A182VKS0 A0A182TZE6 A0A182X960 Q7PSV0 A0A182QFE4 A0A084VIU0 A0A0P4VUN1 R4G4U9 A0A1A9VFS3 A0A182T1J9 A0A182Y4W8 A0A182MA04 A0A0V0G3H4 B4JF35 A0A182IDK5 A0A0M4F4P0 T1PD60 B4PSN4 A0A1W4VMN1 B4QZ72 B4HE39 Q9VFF0 B3P3P8 A0A0A1XFT2 A0A240SWL7 A0A240SXE2 A0A182PL50 B4NJ55 B4M411 A0A182N0I7 U5ETG1 B4K945 A0A067R6C1 A0A182K790 B3LWQ6 A0A1B0FI28 A0A034VJX9 W8B9H4 A0A232EKT3 A0A1W4X8N1 A0A023ESS2 Q17A09 A0A2J7PY28 A0A0P5IWE6 A0A164RZD6 A0A0N8ARF2 A0A0P4ZM21 A0A0N8B320 A0A0P5LT03 A0A0P5FU69 A0A0P5AUN9 A0A0P5K6G8 A0A084VRA3 A0A0P5FZP2 A0A0P5LR24 A0A0P5MNI0 K7IPF7 A0A0C9PY15 E9GEU2 A0A182HLT6 A0A182TZA9 A0A154PGJ5
PDB
3BCC
E-value=1.4385e-93,
Score=876
Ontologies
KEGG
GO
PANTHER
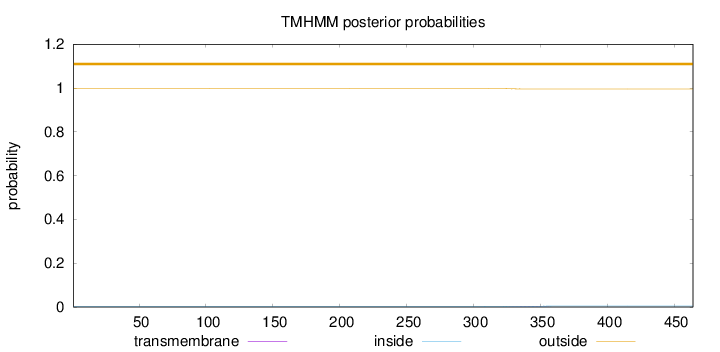
Topology
Length:
464
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03323
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00287
outside
1 - 464
Population Genetic Test Statistics
Pi
13.21837
Theta
15.750477
Tajima's D
-0.513376
CLR
0
CSRT
0.23888805559722
Interpretation
Uncertain
