Gene
KWMTBOMO00023
Pre Gene Modal
BGIBMGA002053
Annotation
PREDICTED:_UDP-sugar_transporter_UST74c_[Amyelois_transitella]
Full name
Ubiquinone biosynthesis protein COQ4 homolog, mitochondrial
+ More
UDP-sugar transporter UST74c
UDP-sugar transporter UST74c
Alternative Name
Coenzyme Q biosynthesis protein 4 homolog
Protein fringe connection
Protein fringe connection
Location in the cell
PlasmaMembrane Reliability : 4.941
Sequence
CDS
ATGTCGGGAAATAGCACACAGGAAGAGGCAGAAGCAGCGGACCGGGCCCAAATGGTCAAAAAAATCCTTAGTGCATTATTTTACGCAATAGCTTCATTCATGATCACGGTTGTCAACAAAACTGTGCTGACTACTTATGCATTCCCTTCATATCAAGTATTAGGCCTGGGACAGATGGTCGCCACCATCATGGTGCTGTGGTTTGGTCGATCAGCCAAACTAGTCCAGTACCCGCCTTTGGATTCAGGCATAGCGCGACGGATATGGCCTCTACCACTGATCTACCTAGGAAACATGGCAACTGGCCTTGGAGGCACCAAAGAATTGAGTTTACCAATGTTCACGGCTCTTCGACGCTTCAGTATTTTGATGACTATGATTTTAGAGCGTATGGTACTTGGTGTAAGAGCATCTTGGCCTGTACAAGCAAGTGTTCTTGCCATGGTAGGAGGTGCTCTGCTTGCTGCTGCTGATGATGTCACCTTCTCTGTTACTGGTTACACCCTTGTACTGCTGAATGATGCATTTACTGCTGCAAATGGTGTATATATGAAGAAAAAGCTCGATGCTAAAGATCTTGGTAAATATGGATTAATGTACTATAATGCACTATTCATGTTTGTCCCAGCAGCATTGGTGGCGTGGAGAACTGGTGACCTTGAGCGATCTGCTGAATATCCTCACTGGAATGATCCATTATTTTTATCACAGTTCTTAATGTCATGCATTATGGGATTTGTTTTGTCTTATAGTGTTATGTTATGTACACAATACAACAGTGCTCTAACCACAACCATAATAGGCTGCTTGAAGAATATCTTGGTCACTTATCTTGGCATGGTAATAGGTGGTGACTATGTTTTCTCCTGGTTGAATTTTGTTGGTTTAAATATCAGCGTATTTGCAAGTCTGTTGTATACTTATGTGACATTCAAGAGAAAACCATCCCCTGCCTATGTGCTTGTGAACAATGGTAAGGTTGATACAGTATAG
Protein
MSGNSTQEEAEAADRAQMVKKILSALFYAIASFMITVVNKTVLTTYAFPSYQVLGLGQMVATIMVLWFGRSAKLVQYPPLDSGIARRIWPLPLIYLGNMATGLGGTKELSLPMFTALRRFSILMTMILERMVLGVRASWPVQASVLAMVGGALLAAADDVTFSVTGYTLVLLNDAFTAANGVYMKKKLDAKDLGKYGLMYYNALFMFVPAALVAWRTGDLERSAEYPHWNDPLFLSQFLMSCIMGFVLSYSVMLCTQYNSALTTTIIGCLKNILVTYLGMVIGGDYVFSWLNFVGLNISVFASLLYTYVTFKRKPSPAYVLVNNGKVDTV
Summary
Description
Component of the coenzyme Q biosynthetic pathway. May play a role in organizing a multi-subunit COQ enzyme complex required for coenzyme Q biosynthesis. Required for steady-state levels of other COQ polypeptides.
Subunit
Component of a multi-subunit COQ enzyme complex.
Similarity
Belongs to the COQ4 family.
Belongs to the TPT transporter family. SLC35D subfamily.
Belongs to the TPT transporter family. SLC35D subfamily.
Keywords
Complete proteome
Golgi apparatus
Membrane
Phosphoprotein
Reference proteome
Sugar transport
Transmembrane
Transmembrane helix
Transport
Feature
chain UDP-sugar transporter UST74c
Uniprot
H9IXS0
A0A2A4JZ73
A0A2H1W0F4
A0A194QY02
A0A194PLP4
A0A212EQF3
+ More
A0A0L7LHV1 N6TZ28 A0A1W4XQ80 D6WEF3 V5GN50 A0A0A1WPI5 F4WYJ8 A0A0K8W6H6 A0A034WEK0 A0A1I8PVA3 B3M6R3 A0A1Y1KRF2 W8C1U5 B4KZ15 E2AQE4 B0WH95 T1PIT7 A0A151WZN5 A0A1W4W3H6 E2C9Q9 B4N6G1 A0A226DQ20 B4QNU9 A0A158P1L5 B4PJV5 A0A195E2S3 A0A1Q3FBS6 B7Z066 Q95YI5 T1H727 A0A195F3C9 Q2LZH8 A0A195D132 A0A151I2Q3 A0A0M4F135 B3NHV4 E9IJM0 R7UXD8 A0A2P8YV15 A0A088A3N6 A0A023ER94 A0A182H0W1 A0A1D2MHG4 A0A3B0JZ14 K7J9N9 A0A0L0BMM1 A0A1B0GPT0 B4LE88 A0A026X4C6 A0A2A3E3Y3 A0A067QNG1 A0A0J7KWA8 Q16FT6 A0A2M4AFE8 A0A154P061 Q16FT7 A0A2M4BT29 A0A1S4G292 B4J253 A0A0L7QS40 A0A0C9R3U7 A0A232F7D9 U5EPZ0 A0A1L8DEB9 A0A3F2Z3G7 A0A1B0FAH2 A0A2P2HXD9 A0A2J7PQK6 A0A1B0AI02 V4A0B0 W5JNW8 A0A2I4BLY5 A0A084WKU7 A0A3B4WZG0 A0A1V4JXM3 A0A0N0U6U1 A0A1B0CRW3 A0A3B3R1M6 A0A0L8HD07 A0A3B3R073 A0A182N1R4 A0A2U9C3R8 A0A336M5X8 A0NDU6 A0A182MKY9 A0A1J1IAK4 A0A2Y9D0Z9 A0A182U1H7 W5MJH7 A0A3B5KIA3 A0A3B4DS94 A7RTQ9 A0A1S3KL08 A0A2Y9D1P7
A0A0L7LHV1 N6TZ28 A0A1W4XQ80 D6WEF3 V5GN50 A0A0A1WPI5 F4WYJ8 A0A0K8W6H6 A0A034WEK0 A0A1I8PVA3 B3M6R3 A0A1Y1KRF2 W8C1U5 B4KZ15 E2AQE4 B0WH95 T1PIT7 A0A151WZN5 A0A1W4W3H6 E2C9Q9 B4N6G1 A0A226DQ20 B4QNU9 A0A158P1L5 B4PJV5 A0A195E2S3 A0A1Q3FBS6 B7Z066 Q95YI5 T1H727 A0A195F3C9 Q2LZH8 A0A195D132 A0A151I2Q3 A0A0M4F135 B3NHV4 E9IJM0 R7UXD8 A0A2P8YV15 A0A088A3N6 A0A023ER94 A0A182H0W1 A0A1D2MHG4 A0A3B0JZ14 K7J9N9 A0A0L0BMM1 A0A1B0GPT0 B4LE88 A0A026X4C6 A0A2A3E3Y3 A0A067QNG1 A0A0J7KWA8 Q16FT6 A0A2M4AFE8 A0A154P061 Q16FT7 A0A2M4BT29 A0A1S4G292 B4J253 A0A0L7QS40 A0A0C9R3U7 A0A232F7D9 U5EPZ0 A0A1L8DEB9 A0A3F2Z3G7 A0A1B0FAH2 A0A2P2HXD9 A0A2J7PQK6 A0A1B0AI02 V4A0B0 W5JNW8 A0A2I4BLY5 A0A084WKU7 A0A3B4WZG0 A0A1V4JXM3 A0A0N0U6U1 A0A1B0CRW3 A0A3B3R1M6 A0A0L8HD07 A0A3B3R073 A0A182N1R4 A0A2U9C3R8 A0A336M5X8 A0NDU6 A0A182MKY9 A0A1J1IAK4 A0A2Y9D0Z9 A0A182U1H7 W5MJH7 A0A3B5KIA3 A0A3B4DS94 A7RTQ9 A0A1S3KL08 A0A2Y9D1P7
Pubmed
19121390
26354079
22118469
26227816
23537049
18362917
+ More
19820115 25830018 21719571 25348373 17994087 28004739 24495485 20798317 25315136 22936249 21347285 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 11533661 12537569 18327897 15632085 21282665 23254933 29403074 24945155 26483478 27289101 20075255 26108605 24508170 24845553 17510324 28648823 20920257 23761445 24438588 29240929 12364791 14747013 17210077 21551351 17615350
19820115 25830018 21719571 25348373 17994087 28004739 24495485 20798317 25315136 22936249 21347285 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 11533661 12537569 18327897 15632085 21282665 23254933 29403074 24945155 26483478 27289101 20075255 26108605 24508170 24845553 17510324 28648823 20920257 23761445 24438588 29240929 12364791 14747013 17210077 21551351 17615350
EMBL
BABH01015629
NWSH01000334
PCG77315.1
ODYU01005594
SOQ46585.1
KQ460953
+ More
KPJ10352.1 KQ459599 KPI94356.1 AGBW02013269 OWR43723.1 JTDY01001079 KOB74939.1 APGK01049656 KB741156 KB630646 KB631725 KB632417 ENN73591.1 ERL83761.1 ERL85609.1 ERL95285.1 KQ971318 EFA00399.1 GALX01005479 JAB62987.1 GBXI01013872 JAD00420.1 GL888450 EGI60730.1 GDHF01031492 GDHF01026034 GDHF01025733 GDHF01021875 GDHF01011003 GDHF01006124 GDHF01005655 JAI20822.1 JAI26280.1 JAI26581.1 JAI30439.1 JAI41311.1 JAI46190.1 JAI46659.1 GAKP01006769 JAC52183.1 CH902618 EDV39749.1 GEZM01075731 JAV63999.1 GAMC01003397 JAC03159.1 CH933809 EDW17812.1 GL441723 EFN64358.1 DS231933 EDS27553.1 KA648020 AFP62649.1 KQ982642 KYQ53246.1 GL453897 EFN75299.1 CH964154 EDW79950.1 LNIX01000014 OXA46944.1 CM000363 CM002912 EDX10851.1 KMZ00229.1 ADTU01006526 CM000159 EDW94724.1 KQ979709 KYN19448.1 GFDL01010097 JAV24948.1 AE014296 BT125707 ACL83317.1 ADO33199.1 AB062677 AY129459 KQ981852 KYN34968.1 CH379069 EAL29529.1 KQ976986 KYN06613.1 KQ976538 KYM81323.1 CP012525 ALC44975.1 CH954178 EDV51969.1 GL763868 EFZ19231.1 AMQN01005894 KB297068 ELU11014.1 PYGN01000344 PSN48088.1 GAPW01002424 JAC11174.1 JXUM01102054 KQ564696 KXJ71880.1 LJIJ01001230 ODM92440.1 OUUW01000012 SPP87315.1 AAZX01008474 JRES01001641 KNC21291.1 AJVK01063842 CH940647 EDW69044.1 KK107024 EZA62254.1 KZ288403 PBC26204.1 KK853201 KDR09907.1 LBMM01002449 KMQ94817.1 CH478382 EAT33099.1 GGFK01006185 MBW39506.1 KQ434783 KZC04734.1 EAT33098.1 GGFJ01007094 MBW56235.1 CH916366 EDV97004.1 KQ414758 KOC61460.1 GBYB01007472 JAG77239.1 NNAY01000786 OXU26512.1 GANO01003511 JAB56360.1 GFDF01009384 JAV04700.1 CCAG010006263 IACF01000487 LAB66256.1 NEVH01022635 PNF18612.1 KB203019 ESO86696.1 ADMH02000800 ETN65003.1 ATLV01024147 KE525350 KFB50841.1 LSYS01005497 OPJ76948.1 KQ435724 KOX78501.1 AJWK01025353 AJWK01025354 AJWK01025355 KQ418483 KOF87176.1 CP026254 AWP11261.1 UFQT01000381 SSX23767.1 AAAB01008900 EAU76830.2 AXCM01001476 CVRI01000043 CRK95990.1 APCN01002820 AHAT01015973 DS469538 EDO45132.1
KPJ10352.1 KQ459599 KPI94356.1 AGBW02013269 OWR43723.1 JTDY01001079 KOB74939.1 APGK01049656 KB741156 KB630646 KB631725 KB632417 ENN73591.1 ERL83761.1 ERL85609.1 ERL95285.1 KQ971318 EFA00399.1 GALX01005479 JAB62987.1 GBXI01013872 JAD00420.1 GL888450 EGI60730.1 GDHF01031492 GDHF01026034 GDHF01025733 GDHF01021875 GDHF01011003 GDHF01006124 GDHF01005655 JAI20822.1 JAI26280.1 JAI26581.1 JAI30439.1 JAI41311.1 JAI46190.1 JAI46659.1 GAKP01006769 JAC52183.1 CH902618 EDV39749.1 GEZM01075731 JAV63999.1 GAMC01003397 JAC03159.1 CH933809 EDW17812.1 GL441723 EFN64358.1 DS231933 EDS27553.1 KA648020 AFP62649.1 KQ982642 KYQ53246.1 GL453897 EFN75299.1 CH964154 EDW79950.1 LNIX01000014 OXA46944.1 CM000363 CM002912 EDX10851.1 KMZ00229.1 ADTU01006526 CM000159 EDW94724.1 KQ979709 KYN19448.1 GFDL01010097 JAV24948.1 AE014296 BT125707 ACL83317.1 ADO33199.1 AB062677 AY129459 KQ981852 KYN34968.1 CH379069 EAL29529.1 KQ976986 KYN06613.1 KQ976538 KYM81323.1 CP012525 ALC44975.1 CH954178 EDV51969.1 GL763868 EFZ19231.1 AMQN01005894 KB297068 ELU11014.1 PYGN01000344 PSN48088.1 GAPW01002424 JAC11174.1 JXUM01102054 KQ564696 KXJ71880.1 LJIJ01001230 ODM92440.1 OUUW01000012 SPP87315.1 AAZX01008474 JRES01001641 KNC21291.1 AJVK01063842 CH940647 EDW69044.1 KK107024 EZA62254.1 KZ288403 PBC26204.1 KK853201 KDR09907.1 LBMM01002449 KMQ94817.1 CH478382 EAT33099.1 GGFK01006185 MBW39506.1 KQ434783 KZC04734.1 EAT33098.1 GGFJ01007094 MBW56235.1 CH916366 EDV97004.1 KQ414758 KOC61460.1 GBYB01007472 JAG77239.1 NNAY01000786 OXU26512.1 GANO01003511 JAB56360.1 GFDF01009384 JAV04700.1 CCAG010006263 IACF01000487 LAB66256.1 NEVH01022635 PNF18612.1 KB203019 ESO86696.1 ADMH02000800 ETN65003.1 ATLV01024147 KE525350 KFB50841.1 LSYS01005497 OPJ76948.1 KQ435724 KOX78501.1 AJWK01025353 AJWK01025354 AJWK01025355 KQ418483 KOF87176.1 CP026254 AWP11261.1 UFQT01000381 SSX23767.1 AAAB01008900 EAU76830.2 AXCM01001476 CVRI01000043 CRK95990.1 APCN01002820 AHAT01015973 DS469538 EDO45132.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000019118 UP000030742 UP000192223 UP000007266 UP000007755 UP000095300 UP000007801 UP000009192 UP000000311 UP000002320 UP000095301 UP000075809 UP000192221 UP000008237 UP000007798 UP000198287 UP000000304 UP000005205 UP000002282 UP000078492 UP000000803 UP000015102 UP000078541 UP000001819 UP000078542 UP000078540 UP000092553 UP000008711 UP000014760 UP000245037 UP000005203 UP000069940 UP000249989 UP000094527 UP000268350 UP000002358 UP000037069 UP000092462 UP000008792 UP000053097 UP000242457 UP000027135 UP000036403 UP000008820 UP000076502 UP000001070 UP000053825 UP000215335 UP000091820 UP000092444 UP000235965 UP000092445 UP000030746 UP000000673 UP000192220 UP000030765 UP000261360 UP000190648 UP000053105 UP000092461 UP000261540 UP000053454 UP000075884 UP000246464 UP000007062 UP000075883 UP000183832 UP000075840 UP000075902 UP000018468 UP000005226 UP000261440 UP000001593 UP000087266 UP000075900
UP000019118 UP000030742 UP000192223 UP000007266 UP000007755 UP000095300 UP000007801 UP000009192 UP000000311 UP000002320 UP000095301 UP000075809 UP000192221 UP000008237 UP000007798 UP000198287 UP000000304 UP000005205 UP000002282 UP000078492 UP000000803 UP000015102 UP000078541 UP000001819 UP000078542 UP000078540 UP000092553 UP000008711 UP000014760 UP000245037 UP000005203 UP000069940 UP000249989 UP000094527 UP000268350 UP000002358 UP000037069 UP000092462 UP000008792 UP000053097 UP000242457 UP000027135 UP000036403 UP000008820 UP000076502 UP000001070 UP000053825 UP000215335 UP000091820 UP000092444 UP000235965 UP000092445 UP000030746 UP000000673 UP000192220 UP000030765 UP000261360 UP000190648 UP000053105 UP000092461 UP000261540 UP000053454 UP000075884 UP000246464 UP000007062 UP000075883 UP000183832 UP000075840 UP000075902 UP000018468 UP000005226 UP000261440 UP000001593 UP000087266 UP000075900
Interpro
SUPFAM
SSF50249
SSF50249
ProteinModelPortal
H9IXS0
A0A2A4JZ73
A0A2H1W0F4
A0A194QY02
A0A194PLP4
A0A212EQF3
+ More
A0A0L7LHV1 N6TZ28 A0A1W4XQ80 D6WEF3 V5GN50 A0A0A1WPI5 F4WYJ8 A0A0K8W6H6 A0A034WEK0 A0A1I8PVA3 B3M6R3 A0A1Y1KRF2 W8C1U5 B4KZ15 E2AQE4 B0WH95 T1PIT7 A0A151WZN5 A0A1W4W3H6 E2C9Q9 B4N6G1 A0A226DQ20 B4QNU9 A0A158P1L5 B4PJV5 A0A195E2S3 A0A1Q3FBS6 B7Z066 Q95YI5 T1H727 A0A195F3C9 Q2LZH8 A0A195D132 A0A151I2Q3 A0A0M4F135 B3NHV4 E9IJM0 R7UXD8 A0A2P8YV15 A0A088A3N6 A0A023ER94 A0A182H0W1 A0A1D2MHG4 A0A3B0JZ14 K7J9N9 A0A0L0BMM1 A0A1B0GPT0 B4LE88 A0A026X4C6 A0A2A3E3Y3 A0A067QNG1 A0A0J7KWA8 Q16FT6 A0A2M4AFE8 A0A154P061 Q16FT7 A0A2M4BT29 A0A1S4G292 B4J253 A0A0L7QS40 A0A0C9R3U7 A0A232F7D9 U5EPZ0 A0A1L8DEB9 A0A3F2Z3G7 A0A1B0FAH2 A0A2P2HXD9 A0A2J7PQK6 A0A1B0AI02 V4A0B0 W5JNW8 A0A2I4BLY5 A0A084WKU7 A0A3B4WZG0 A0A1V4JXM3 A0A0N0U6U1 A0A1B0CRW3 A0A3B3R1M6 A0A0L8HD07 A0A3B3R073 A0A182N1R4 A0A2U9C3R8 A0A336M5X8 A0NDU6 A0A182MKY9 A0A1J1IAK4 A0A2Y9D0Z9 A0A182U1H7 W5MJH7 A0A3B5KIA3 A0A3B4DS94 A7RTQ9 A0A1S3KL08 A0A2Y9D1P7
A0A0L7LHV1 N6TZ28 A0A1W4XQ80 D6WEF3 V5GN50 A0A0A1WPI5 F4WYJ8 A0A0K8W6H6 A0A034WEK0 A0A1I8PVA3 B3M6R3 A0A1Y1KRF2 W8C1U5 B4KZ15 E2AQE4 B0WH95 T1PIT7 A0A151WZN5 A0A1W4W3H6 E2C9Q9 B4N6G1 A0A226DQ20 B4QNU9 A0A158P1L5 B4PJV5 A0A195E2S3 A0A1Q3FBS6 B7Z066 Q95YI5 T1H727 A0A195F3C9 Q2LZH8 A0A195D132 A0A151I2Q3 A0A0M4F135 B3NHV4 E9IJM0 R7UXD8 A0A2P8YV15 A0A088A3N6 A0A023ER94 A0A182H0W1 A0A1D2MHG4 A0A3B0JZ14 K7J9N9 A0A0L0BMM1 A0A1B0GPT0 B4LE88 A0A026X4C6 A0A2A3E3Y3 A0A067QNG1 A0A0J7KWA8 Q16FT6 A0A2M4AFE8 A0A154P061 Q16FT7 A0A2M4BT29 A0A1S4G292 B4J253 A0A0L7QS40 A0A0C9R3U7 A0A232F7D9 U5EPZ0 A0A1L8DEB9 A0A3F2Z3G7 A0A1B0FAH2 A0A2P2HXD9 A0A2J7PQK6 A0A1B0AI02 V4A0B0 W5JNW8 A0A2I4BLY5 A0A084WKU7 A0A3B4WZG0 A0A1V4JXM3 A0A0N0U6U1 A0A1B0CRW3 A0A3B3R1M6 A0A0L8HD07 A0A3B3R073 A0A182N1R4 A0A2U9C3R8 A0A336M5X8 A0NDU6 A0A182MKY9 A0A1J1IAK4 A0A2Y9D0Z9 A0A182U1H7 W5MJH7 A0A3B5KIA3 A0A3B4DS94 A7RTQ9 A0A1S3KL08 A0A2Y9D1P7
PDB
5OGK
E-value=0.00537119,
Score=93
Ontologies
GO
GO:0016021
GO:0008643
GO:0005462
GO:0015165
GO:0022857
GO:0005463
GO:0005794
GO:0005461
GO:0015297
GO:0005464
GO:0007447
GO:0005797
GO:0007476
GO:0005459
GO:0007367
GO:0005460
GO:0001745
GO:0006744
GO:0031314
GO:0015790
GO:0000139
GO:0009880
GO:1990570
GO:0015786
GO:0015787
GO:0007423
GO:0015783
GO:0072334
GO:1990569
GO:0005743
Topology
Subcellular location
Mitochondrion inner membrane
Golgi apparatus membrane
Golgi apparatus membrane
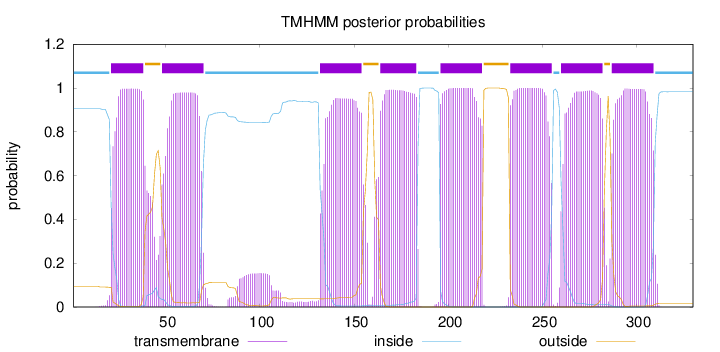
Length:
330
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
176.08256
Exp number, first 60 AAs:
31.91792
Total prob of N-in:
0.90539
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 38
outside
39 - 47
TMhelix
48 - 70
inside
71 - 131
TMhelix
132 - 154
outside
155 - 163
TMhelix
164 - 183
inside
184 - 195
TMhelix
196 - 218
outside
219 - 232
TMhelix
233 - 255
inside
256 - 259
TMhelix
260 - 282
outside
283 - 286
TMhelix
287 - 309
inside
310 - 330
Population Genetic Test Statistics
Pi
6.43936
Theta
6.051125
Tajima's D
0.18046
CLR
0.723097
CSRT
0.439978001099945
Interpretation
Uncertain
