Gene
KWMTBOMO00022
Pre Gene Modal
BGIBMGA002079
Annotation
PREDICTED:_ephrin_type-B_receptor_1-B_[Plutella_xylostella]
Full name
Receptor protein-tyrosine kinase
Location in the cell
PlasmaMembrane Reliability : 1.858
Sequence
CDS
ATGATTTGGCTGATAGTAGTGACGGCCTTGGCTAGCGTGCATGGCGAACAAGTGCTGCTTCTCGACACTACAGCAGAAGATACTCTAGGCTGGACCCGGTTCTCGTATGGTGCGCAGGCTAGCACTGCCGGCTGGCTCGAAGAATCTTACACTAACTTTGAAAAACAAATAAACTGGCGCTCCTATGTGGTCTGTGACGTCGCCCACCATAATGTGAATAACTGGCTCTGGACCCCGTTCATCGAACGCAGAAATGCCAATAGAATCTACATAGAAATCAAGTTTACTATTCGTGATTGCTCACTTTTCCCAGGAAATGCATTGAGTTGTAAAGAAACATTCAGTCTCTTGTATTATGAATTTGATGTTGCCACTAGAGAACAACCTCCCTGGGAGCCCGAGAGTTACAAGTTAGTTGGCCGCATCGCCGCAGGGGAAGGCAGATTTAACACTAACTCTGAAGTGGACATCAACACTGAAGTGAAGAGTATTGCAGTCACAAAGAAAGGTGTGTATTTTGCATTTAGAGATCAAGGAGCCTGCATATCTATTTTGGCAGTGAAAGTCTATTACATCACATGTCCTGATGTCAACATTAACTTTGCAAGATTCCCTGCCACTCCCACTGGCAGGGAAGTTACGGTTATTGAACAAGCCAATGGAACTTGTGTTGCTAATGCTGAGGTTGCTCCTGGTGAAAAAGCCCCTGTTTATTTATGCAAGGGTGATGGCAAATGGACCCTTCCCAGTGGCTCTTGCAAATGCAGGGCAGGCTTTGAACCTGACCACAACAATCAAATTTGTACAGAATGTGCACCTGGAAAATTTAAATCTCACACTGGTGACGAACACTGCATCCCGTGCCCTGATCATTCTGAGTCAACCATATATGCTGCTGCTGAATGTAAATGTACTCCAAAATACTATAGAGCTAAGAAAGACCCAAAAACCATACCCTGTACTCAACCACCTTCAGCACCTGATAATTTGACTGTAACATTTGCTGATCAGTTTTCTATTTCTCTTACTTGGCAACCGCCACATAATACTGGCGGAAGAAGTGATATTACATATAAGATTTCATGTGCCAATTGCGGCCCCACTGTGGAATTTAGACCAGGAGAGACTGGCCTTAATAGTACACGAGTTACAATTAGTGGTCTAGATCCAGTAACTGAATACAAGTTTCAAGTTTATGCAGAAAATGGAGTATCTGAACTCTCTCCTGATCCTGCAAAACACATAGAAATCACAGCAGTAACTGAAGCCTCTGTAGTGAGTGTAGTGTCAAAATTGAGGATTTTGAACACAGAAAGTGATAGGTTAACAATTGCTTGGAATCCACCACAAGTAGACTTAACTGATCCTGATGACACTATAGAGAGCTATGAAGTGAAATGCTTTCCAAAAGACAGTCAAGACAAGAATACCAACACCACCCTGAGGATTACTAAGGACCCACAGGTCACCATTACAGGATTAAGAAAAGACACTGAATATGGCATTAGGGTCAGAGCCAAGATGAAGAGAGGATGGGGAGAATTCAGTGGAATTGTATATGCTCGAACCAATTCTGTTACTGAAACTTCATTCATTGACGAAGAAGGAGCTCAAGTTAGATTAGTTGCTGGTGTTATGGTGGCAGTAGTTGTCCTCACAGTGATTGCCATCATTGCTACTGTTTTATTTTTGAGATCTCGTGCTGATGATGAATGTGATAAAAAACAACCTAGTGATTGTAATGCTCTTAATTATAGAAATGGAGAAGTTTACACAGGACCTGATCGTCCTGCTAAGACAAGCAGCAATGCTACCACTCCTCTTTTTGCTGGCACTGGTTCCAGGACTTACATAGACCCTCATACATATGAAGACCCCAATCAAGCTGTCAGAGAATTTGCACGTGAGATTGATGCATCATGTATTACTATTGAAGCAATAATAGGTGGTGGTGAATTTGGAGATGTTTGTCGTGGCAAATTGAAACTTCCAGCTAGTTGTGGCCCAGAAATTGATGTTGCCATCAAGACATTAAAACCAGGCTCCACTGAACGAGCTCGACGTGATTTCCTGGCTGAAGCGTCTATAATGGGCCAATTTGAACATCCAAATGTGATATTCTTACAGGGAGTCGTCACAAAATGCAATCCAATCATGATTATTACTGAGTTCATGGAAAATGGATCTTTGGACACTTTCTTACGGGCGAATGATGGAAAATTTAGAGTGCTGCAATTGGTTGGTATGTTACGTGGTATCGCCACTGGAATGCAATATCTTTCAGAGATGAACTACATCCATCGTGACCTAGCTGCACGAAATGTACTTGTCAATTCTCAACTTGTTTGTAAGATAGCTGACTTTGGTCTCTCTCGTGAAATTGAATCTACTGCTGATGGAGCCTATACAACCCGTGGTGGAAAGATTCCAGTGCGATGGACAGCACCAGAAGCAATAGCATTTAGGAAATTTACTTCTGCCAGTGATGTCTGGTCTATGGGAATAGTGTGCTGGGAGGTAATGAGTTTTGGGGAACGACCTTATTGGAACTGGAGCAACCAAGATGTGATTAAATCAATTGAAAAAGGATATCGTCTACCAGCTCCTATGGACTGTCCTGAGGCTGTGTATCAGCTGATGCTTGATTGCTGGCAGAAAGAACGAACACACAGACCAACTTTTTCAAGCATTGTGAAGACACTAGATAAATTAATTCGTTGCCCAGATACATTGAGAAAGATTGCCCAAAACCGTTCAGCAGACCCTTTAGCAGGCGATACTCCAGACTTGATTCAGTTTACATCAGTGGAAGAATGGCTTGAATGCATAAAAATGTCACGGTACATCGAGAAGTTTCGAGCGGCGGGCATTTCCGACATGGATGCGGTGGTCGACTTGACTGTGCATCAGCTGGCATCTCTCGGCGTGACTCTTGTCGGTCATCAGAAGAAGATTATGAATAGTGTGCAGAGCATGCGGGCTCAGATCCGGGGCAGCGGTCCTTACGGATTTCTTGTATAG
Protein
MIWLIVVTALASVHGEQVLLLDTTAEDTLGWTRFSYGAQASTAGWLEESYTNFEKQINWRSYVVCDVAHHNVNNWLWTPFIERRNANRIYIEIKFTIRDCSLFPGNALSCKETFSLLYYEFDVATREQPPWEPESYKLVGRIAAGEGRFNTNSEVDINTEVKSIAVTKKGVYFAFRDQGACISILAVKVYYITCPDVNINFARFPATPTGREVTVIEQANGTCVANAEVAPGEKAPVYLCKGDGKWTLPSGSCKCRAGFEPDHNNQICTECAPGKFKSHTGDEHCIPCPDHSESTIYAAAECKCTPKYYRAKKDPKTIPCTQPPSAPDNLTVTFADQFSISLTWQPPHNTGGRSDITYKISCANCGPTVEFRPGETGLNSTRVTISGLDPVTEYKFQVYAENGVSELSPDPAKHIEITAVTEASVVSVVSKLRILNTESDRLTIAWNPPQVDLTDPDDTIESYEVKCFPKDSQDKNTNTTLRITKDPQVTITGLRKDTEYGIRVRAKMKRGWGEFSGIVYARTNSVTETSFIDEEGAQVRLVAGVMVAVVVLTVIAIIATVLFLRSRADDECDKKQPSDCNALNYRNGEVYTGPDRPAKTSSNATTPLFAGTGSRTYIDPHTYEDPNQAVREFAREIDASCITIEAIIGGGEFGDVCRGKLKLPASCGPEIDVAIKTLKPGSTERARRDFLAEASIMGQFEHPNVIFLQGVVTKCNPIMIITEFMENGSLDTFLRANDGKFRVLQLVGMLRGIATGMQYLSEMNYIHRDLAARNVLVNSQLVCKIADFGLSREIESTADGAYTTRGGKIPVRWTAPEAIAFRKFTSASDVWSMGIVCWEVMSFGERPYWNWSNQDVIKSIEKGYRLPAPMDCPEAVYQLMLDCWQKERTHRPTFSSIVKTLDKLIRCPDTLRKIAQNRSADPLAGDTPDLIQFTSVEEWLECIKMSRYIEKFRAAGISDMDAVVDLTVHQLASLGVTLVGHQKKIMNSVQSMRAQIRGSGPYGFLV
Summary
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Feature
chain Receptor protein-tyrosine kinase
Uniprot
A0A2A4JJG1
Q6VU50
A0A194PT94
A0A0L7LI74
A0A194R3D3
H9IXU6
+ More
A0A1B6MMJ9 A0A1Y1NDI2 A0A1B6KV88 A0A0A9XH54 A0A146KLG2 A0A146LRQ2 A0A146KR58 A0A1W4X9Y2 A0A1W4WZ35 A0A0C9PGV0 X1WQ64 A0A3L8E358 A0A158P3E8 Q5D184 A0A151WZE6 A0A2S2PM87 A0A195EZ24 K7IQA3 T1I7Z0 A0A0M9AAF8 A0A2A3ENR8 A0A0P5C6L9 E0W1E4 A0A0N8E0W6 A0A0P4ZRK1 A0A139WE15 A0A0J7L2X8 A0A0P5P411 A0A0P5E3K1 A0A0P5CF79 A0A0P5J8X4 A0A0P5DAF3 A0A0P4ZVS8 A0A0P4XL12 A0A0P4ZR96 A0A0P5N8Z4 A0A0P5GG86 A0A0N8AHC6 A0A0P5YDB9 A0A0P5NAX6 A0A0P6I4L8 A0A0P5XDR9 A0A0P6C1D6 A0A0P5AJG0 A0A1W4WZ30 A0A1W4WZ71 A0A1W4X8H2 A0A0C9PGV6 E9GM17 A0A1W4X9R7 E2C3J5 A0A087ZS47 A0A147BCU0 A0A026W547 A0A0L7RCX9 A0A195BIF7 A0A226E4M0 E9IIL2 A0A0P5Y6S7 A0A0P5BE05 A0A195CCU3 A0A0P5IAT1 A0A0P4Z8N5 A0A0P5VQD8 A0A195DN87 A0A0P6A272 A0A0P5QSN2 A0A0N8A7B5 A0A0N8CXX1 A0A0P4XJK5 A0A0P5IVR6 A0A0P6GDT6 A0A0P6CXK8 A0A0P4XMR5 A0A154P9Y2 T1L496 A0A2J7QUT8 A0A3B0K475 B4H885 B4L7C3 A0A034VY46 B4MF10 D0Z7B1 A0A0A1WQS3 W8BRA0 A0A0L0CNY5 A0A3B0K6U5 A0A0K8TYV5 W8CD17 B7QJ81 B3P9S4 B4PW17 A0A1W4VIH8 A0A0Q9XFB8
A0A1B6MMJ9 A0A1Y1NDI2 A0A1B6KV88 A0A0A9XH54 A0A146KLG2 A0A146LRQ2 A0A146KR58 A0A1W4X9Y2 A0A1W4WZ35 A0A0C9PGV0 X1WQ64 A0A3L8E358 A0A158P3E8 Q5D184 A0A151WZE6 A0A2S2PM87 A0A195EZ24 K7IQA3 T1I7Z0 A0A0M9AAF8 A0A2A3ENR8 A0A0P5C6L9 E0W1E4 A0A0N8E0W6 A0A0P4ZRK1 A0A139WE15 A0A0J7L2X8 A0A0P5P411 A0A0P5E3K1 A0A0P5CF79 A0A0P5J8X4 A0A0P5DAF3 A0A0P4ZVS8 A0A0P4XL12 A0A0P4ZR96 A0A0P5N8Z4 A0A0P5GG86 A0A0N8AHC6 A0A0P5YDB9 A0A0P5NAX6 A0A0P6I4L8 A0A0P5XDR9 A0A0P6C1D6 A0A0P5AJG0 A0A1W4WZ30 A0A1W4WZ71 A0A1W4X8H2 A0A0C9PGV6 E9GM17 A0A1W4X9R7 E2C3J5 A0A087ZS47 A0A147BCU0 A0A026W547 A0A0L7RCX9 A0A195BIF7 A0A226E4M0 E9IIL2 A0A0P5Y6S7 A0A0P5BE05 A0A195CCU3 A0A0P5IAT1 A0A0P4Z8N5 A0A0P5VQD8 A0A195DN87 A0A0P6A272 A0A0P5QSN2 A0A0N8A7B5 A0A0N8CXX1 A0A0P4XJK5 A0A0P5IVR6 A0A0P6GDT6 A0A0P6CXK8 A0A0P4XMR5 A0A154P9Y2 T1L496 A0A2J7QUT8 A0A3B0K475 B4H885 B4L7C3 A0A034VY46 B4MF10 D0Z7B1 A0A0A1WQS3 W8BRA0 A0A0L0CNY5 A0A3B0K6U5 A0A0K8TYV5 W8CD17 B7QJ81 B3P9S4 B4PW17 A0A1W4VIH8 A0A0Q9XFB8
EC Number
2.7.10.1
Pubmed
EMBL
NWSH01001255
PCG71969.1
AY327249
AAQ67231.1
KQ459599
KPI94355.1
+ More
JTDY01001079 KOB74936.1 KQ460953 KPJ10351.1 BABH01015624 BABH01015625 GEBQ01002844 JAT37133.1 GEZM01007759 GEZM01007758 JAV94950.1 GEBQ01024618 JAT15359.1 GBHO01023552 JAG20052.1 GDHC01021225 JAP97403.1 GDHC01009529 JAQ09100.1 GDHC01019741 JAP98887.1 GBYB01000098 JAG69865.1 ABLF02038853 ABLF02038859 ABLF02038862 ABLF02038869 QOIP01000001 RLU26962.1 ADTU01007737 ADTU01007738 ADTU01007739 ADTU01007740 ADTU01007741 ADTU01007742 ADTU01007743 ADTU01007744 AY921579 AAX14899.1 KQ982649 KYQ53071.1 GGMR01017885 MBY30504.1 KQ981905 KYN33555.1 ACPB03012482 ACPB03012483 ACPB03012484 KQ435714 KOX79159.1 KZ288204 PBC33154.1 GDIP01175125 GDIQ01036299 JAJ48277.1 JAN58438.1 DS235870 EEB19450.1 GDIQ01072740 JAN21997.1 GDIP01209014 JAJ14388.1 KQ971357 KYB26081.1 LBMM01001046 KMQ97021.1 GDIQ01154805 JAK96920.1 GDIP01147330 GDIQ01160807 JAJ76072.1 JAK90918.1 GDIP01175126 JAJ48276.1 GDIQ01209341 JAK42384.1 GDIP01175127 GDIQ01184451 JAJ48275.1 JAK67274.1 GDIP01209012 JAJ14390.1 GDIP01240374 JAI83027.1 GDIP01209013 JAJ14389.1 GDIQ01148266 JAL03460.1 GDIP01147328 GDIQ01242824 JAJ76074.1 JAK08901.1 GDIP01147329 JAJ76073.1 GDIP01073645 JAM30070.1 GDIQ01148265 JAL03461.1 GDIQ01009418 JAN85319.1 GDIP01073644 JAM30071.1 GDIP01010921 JAM92794.1 GDIP01198125 JAJ25277.1 GBYB01000103 JAG69870.1 GL732552 EFX79285.1 GL452320 EFN77533.1 GEGO01006835 JAR88569.1 KK107447 EZA50746.1 KQ414615 KOC68645.1 KQ976467 KYM84152.1 LNIX01000007 OXA51974.1 GL763494 EFZ19580.1 GDIQ01244028 GDIP01063745 JAK07697.1 JAM39970.1 GDIP01185868 JAJ37534.1 KQ977935 KYM98679.1 GDIQ01239879 JAK11846.1 GDIP01217917 GDIQ01029110 JAJ05485.1 JAN65627.1 GDIP01096885 JAM06830.1 KQ980713 KYN14291.1 GDIQ01132673 GDIP01036243 JAL19053.1 JAM67472.1 GDIQ01110127 JAL41599.1 GDIP01175417 JAJ47985.1 GDIP01088079 GDIQ01084635 JAM15636.1 JAN10102.1 GDIP01240373 JAI83028.1 GDIQ01209340 JAK42385.1 GDIQ01036298 JAN58439.1 GDIQ01086187 JAN08550.1 GDIP01240372 JAI83029.1 KQ434853 KZC08637.1 CAEY01001080 NEVH01010480 PNF32350.1 OUUW01000017 SPP89004.1 CH479221 EDW34885.1 CH933813 EDW10917.2 GAKP01010751 JAC48201.1 CH940665 EDW71111.2 KRF85367.1 CM000831 ACY70479.1 GBXI01013291 JAD01001.1 GAMC01002725 JAC03831.1 JRES01000108 KNC34063.1 SPP89003.1 GDHF01032667 GDHF01027214 GDHF01017643 GDHF01000863 JAI19647.1 JAI25100.1 JAI34671.1 JAI51451.1 GAMC01002727 GAMC01002726 JAC03830.1 ABJB010182879 ABJB010388182 ABJB010401843 ABJB010475459 ABJB010801537 ABJB010836030 ABJB010973941 DS950795 EEC18904.1 CH954184 EDV45237.1 CM000161 EDW99322.2 KRK04916.1 KRG07276.1
JTDY01001079 KOB74936.1 KQ460953 KPJ10351.1 BABH01015624 BABH01015625 GEBQ01002844 JAT37133.1 GEZM01007759 GEZM01007758 JAV94950.1 GEBQ01024618 JAT15359.1 GBHO01023552 JAG20052.1 GDHC01021225 JAP97403.1 GDHC01009529 JAQ09100.1 GDHC01019741 JAP98887.1 GBYB01000098 JAG69865.1 ABLF02038853 ABLF02038859 ABLF02038862 ABLF02038869 QOIP01000001 RLU26962.1 ADTU01007737 ADTU01007738 ADTU01007739 ADTU01007740 ADTU01007741 ADTU01007742 ADTU01007743 ADTU01007744 AY921579 AAX14899.1 KQ982649 KYQ53071.1 GGMR01017885 MBY30504.1 KQ981905 KYN33555.1 ACPB03012482 ACPB03012483 ACPB03012484 KQ435714 KOX79159.1 KZ288204 PBC33154.1 GDIP01175125 GDIQ01036299 JAJ48277.1 JAN58438.1 DS235870 EEB19450.1 GDIQ01072740 JAN21997.1 GDIP01209014 JAJ14388.1 KQ971357 KYB26081.1 LBMM01001046 KMQ97021.1 GDIQ01154805 JAK96920.1 GDIP01147330 GDIQ01160807 JAJ76072.1 JAK90918.1 GDIP01175126 JAJ48276.1 GDIQ01209341 JAK42384.1 GDIP01175127 GDIQ01184451 JAJ48275.1 JAK67274.1 GDIP01209012 JAJ14390.1 GDIP01240374 JAI83027.1 GDIP01209013 JAJ14389.1 GDIQ01148266 JAL03460.1 GDIP01147328 GDIQ01242824 JAJ76074.1 JAK08901.1 GDIP01147329 JAJ76073.1 GDIP01073645 JAM30070.1 GDIQ01148265 JAL03461.1 GDIQ01009418 JAN85319.1 GDIP01073644 JAM30071.1 GDIP01010921 JAM92794.1 GDIP01198125 JAJ25277.1 GBYB01000103 JAG69870.1 GL732552 EFX79285.1 GL452320 EFN77533.1 GEGO01006835 JAR88569.1 KK107447 EZA50746.1 KQ414615 KOC68645.1 KQ976467 KYM84152.1 LNIX01000007 OXA51974.1 GL763494 EFZ19580.1 GDIQ01244028 GDIP01063745 JAK07697.1 JAM39970.1 GDIP01185868 JAJ37534.1 KQ977935 KYM98679.1 GDIQ01239879 JAK11846.1 GDIP01217917 GDIQ01029110 JAJ05485.1 JAN65627.1 GDIP01096885 JAM06830.1 KQ980713 KYN14291.1 GDIQ01132673 GDIP01036243 JAL19053.1 JAM67472.1 GDIQ01110127 JAL41599.1 GDIP01175417 JAJ47985.1 GDIP01088079 GDIQ01084635 JAM15636.1 JAN10102.1 GDIP01240373 JAI83028.1 GDIQ01209340 JAK42385.1 GDIQ01036298 JAN58439.1 GDIQ01086187 JAN08550.1 GDIP01240372 JAI83029.1 KQ434853 KZC08637.1 CAEY01001080 NEVH01010480 PNF32350.1 OUUW01000017 SPP89004.1 CH479221 EDW34885.1 CH933813 EDW10917.2 GAKP01010751 JAC48201.1 CH940665 EDW71111.2 KRF85367.1 CM000831 ACY70479.1 GBXI01013291 JAD01001.1 GAMC01002725 JAC03831.1 JRES01000108 KNC34063.1 SPP89003.1 GDHF01032667 GDHF01027214 GDHF01017643 GDHF01000863 JAI19647.1 JAI25100.1 JAI34671.1 JAI51451.1 GAMC01002727 GAMC01002726 JAC03830.1 ABJB010182879 ABJB010388182 ABJB010401843 ABJB010475459 ABJB010801537 ABJB010836030 ABJB010973941 DS950795 EEC18904.1 CH954184 EDV45237.1 CM000161 EDW99322.2 KRK04916.1 KRG07276.1
Proteomes
UP000218220
UP000053268
UP000037510
UP000053240
UP000005204
UP000192223
+ More
UP000007819 UP000279307 UP000005205 UP000075809 UP000078541 UP000002358 UP000015103 UP000053105 UP000242457 UP000009046 UP000007266 UP000036403 UP000000305 UP000008237 UP000005203 UP000053097 UP000053825 UP000078540 UP000198287 UP000078542 UP000078492 UP000076502 UP000015104 UP000235965 UP000268350 UP000008744 UP000009192 UP000008792 UP000037069 UP000001555 UP000008711 UP000002282 UP000192221
UP000007819 UP000279307 UP000005205 UP000075809 UP000078541 UP000002358 UP000015103 UP000053105 UP000242457 UP000009046 UP000007266 UP000036403 UP000000305 UP000008237 UP000005203 UP000053097 UP000053825 UP000078540 UP000198287 UP000078542 UP000078492 UP000076502 UP000015104 UP000235965 UP000268350 UP000008744 UP000009192 UP000008792 UP000037069 UP000001555 UP000008711 UP000002282 UP000192221
Pfam
Interpro
IPR011009
Kinase-like_dom_sf
+ More
IPR016257 Tyr_kinase_ephrin_rcpt
IPR001090 Ephrin_rcpt_lig-bd_dom
IPR036116 FN3_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR001660 SAM
IPR003961 FN3_dom
IPR027936 Eph_TM
IPR009030 Growth_fac_rcpt_cys_sf
IPR017441 Protein_kinase_ATP_BS
IPR013783 Ig-like_fold
IPR013761 SAM/pointed_sf
IPR008979 Galactose-bd-like_sf
IPR020635 Tyr_kinase_cat_dom
IPR008266 Tyr_kinase_AS
IPR001426 Tyr_kinase_rcpt_V_CS
IPR000719 Prot_kinase_dom
IPR011641 Tyr-kin_ephrin_A/B_rcpt-like
IPR016257 Tyr_kinase_ephrin_rcpt
IPR001090 Ephrin_rcpt_lig-bd_dom
IPR036116 FN3_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR001660 SAM
IPR003961 FN3_dom
IPR027936 Eph_TM
IPR009030 Growth_fac_rcpt_cys_sf
IPR017441 Protein_kinase_ATP_BS
IPR013783 Ig-like_fold
IPR013761 SAM/pointed_sf
IPR008979 Galactose-bd-like_sf
IPR020635 Tyr_kinase_cat_dom
IPR008266 Tyr_kinase_AS
IPR001426 Tyr_kinase_rcpt_V_CS
IPR000719 Prot_kinase_dom
IPR011641 Tyr-kin_ephrin_A/B_rcpt-like
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A2A4JJG1
Q6VU50
A0A194PT94
A0A0L7LI74
A0A194R3D3
H9IXU6
+ More
A0A1B6MMJ9 A0A1Y1NDI2 A0A1B6KV88 A0A0A9XH54 A0A146KLG2 A0A146LRQ2 A0A146KR58 A0A1W4X9Y2 A0A1W4WZ35 A0A0C9PGV0 X1WQ64 A0A3L8E358 A0A158P3E8 Q5D184 A0A151WZE6 A0A2S2PM87 A0A195EZ24 K7IQA3 T1I7Z0 A0A0M9AAF8 A0A2A3ENR8 A0A0P5C6L9 E0W1E4 A0A0N8E0W6 A0A0P4ZRK1 A0A139WE15 A0A0J7L2X8 A0A0P5P411 A0A0P5E3K1 A0A0P5CF79 A0A0P5J8X4 A0A0P5DAF3 A0A0P4ZVS8 A0A0P4XL12 A0A0P4ZR96 A0A0P5N8Z4 A0A0P5GG86 A0A0N8AHC6 A0A0P5YDB9 A0A0P5NAX6 A0A0P6I4L8 A0A0P5XDR9 A0A0P6C1D6 A0A0P5AJG0 A0A1W4WZ30 A0A1W4WZ71 A0A1W4X8H2 A0A0C9PGV6 E9GM17 A0A1W4X9R7 E2C3J5 A0A087ZS47 A0A147BCU0 A0A026W547 A0A0L7RCX9 A0A195BIF7 A0A226E4M0 E9IIL2 A0A0P5Y6S7 A0A0P5BE05 A0A195CCU3 A0A0P5IAT1 A0A0P4Z8N5 A0A0P5VQD8 A0A195DN87 A0A0P6A272 A0A0P5QSN2 A0A0N8A7B5 A0A0N8CXX1 A0A0P4XJK5 A0A0P5IVR6 A0A0P6GDT6 A0A0P6CXK8 A0A0P4XMR5 A0A154P9Y2 T1L496 A0A2J7QUT8 A0A3B0K475 B4H885 B4L7C3 A0A034VY46 B4MF10 D0Z7B1 A0A0A1WQS3 W8BRA0 A0A0L0CNY5 A0A3B0K6U5 A0A0K8TYV5 W8CD17 B7QJ81 B3P9S4 B4PW17 A0A1W4VIH8 A0A0Q9XFB8
A0A1B6MMJ9 A0A1Y1NDI2 A0A1B6KV88 A0A0A9XH54 A0A146KLG2 A0A146LRQ2 A0A146KR58 A0A1W4X9Y2 A0A1W4WZ35 A0A0C9PGV0 X1WQ64 A0A3L8E358 A0A158P3E8 Q5D184 A0A151WZE6 A0A2S2PM87 A0A195EZ24 K7IQA3 T1I7Z0 A0A0M9AAF8 A0A2A3ENR8 A0A0P5C6L9 E0W1E4 A0A0N8E0W6 A0A0P4ZRK1 A0A139WE15 A0A0J7L2X8 A0A0P5P411 A0A0P5E3K1 A0A0P5CF79 A0A0P5J8X4 A0A0P5DAF3 A0A0P4ZVS8 A0A0P4XL12 A0A0P4ZR96 A0A0P5N8Z4 A0A0P5GG86 A0A0N8AHC6 A0A0P5YDB9 A0A0P5NAX6 A0A0P6I4L8 A0A0P5XDR9 A0A0P6C1D6 A0A0P5AJG0 A0A1W4WZ30 A0A1W4WZ71 A0A1W4X8H2 A0A0C9PGV6 E9GM17 A0A1W4X9R7 E2C3J5 A0A087ZS47 A0A147BCU0 A0A026W547 A0A0L7RCX9 A0A195BIF7 A0A226E4M0 E9IIL2 A0A0P5Y6S7 A0A0P5BE05 A0A195CCU3 A0A0P5IAT1 A0A0P4Z8N5 A0A0P5VQD8 A0A195DN87 A0A0P6A272 A0A0P5QSN2 A0A0N8A7B5 A0A0N8CXX1 A0A0P4XJK5 A0A0P5IVR6 A0A0P6GDT6 A0A0P6CXK8 A0A0P4XMR5 A0A154P9Y2 T1L496 A0A2J7QUT8 A0A3B0K475 B4H885 B4L7C3 A0A034VY46 B4MF10 D0Z7B1 A0A0A1WQS3 W8BRA0 A0A0L0CNY5 A0A3B0K6U5 A0A0K8TYV5 W8CD17 B7QJ81 B3P9S4 B4PW17 A0A1W4VIH8 A0A0Q9XFB8
PDB
2HEL
E-value=4.827e-137,
Score=1254
Ontologies
GO
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.994029
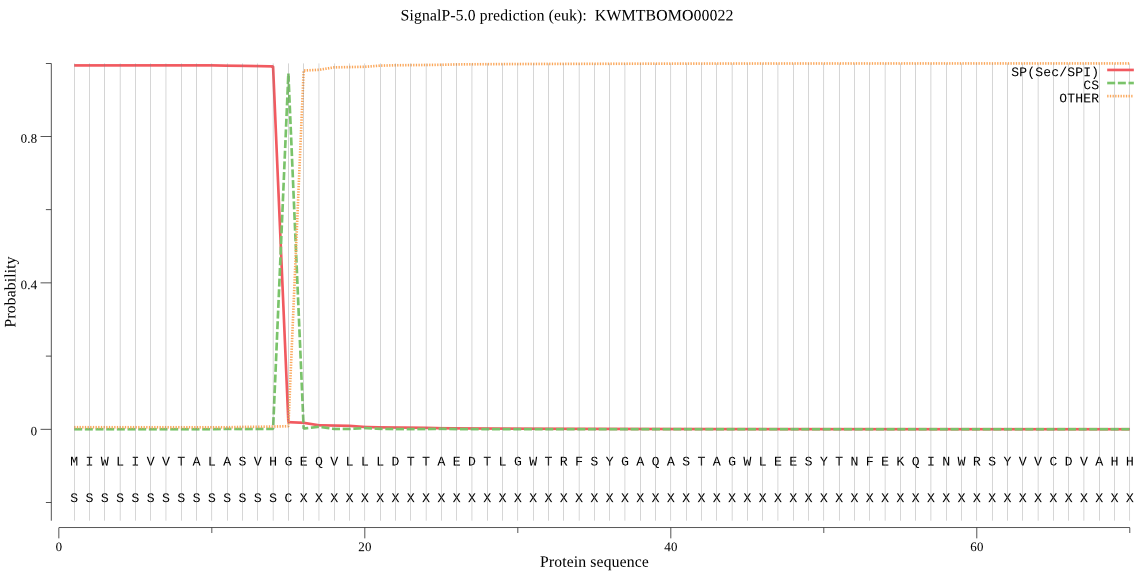
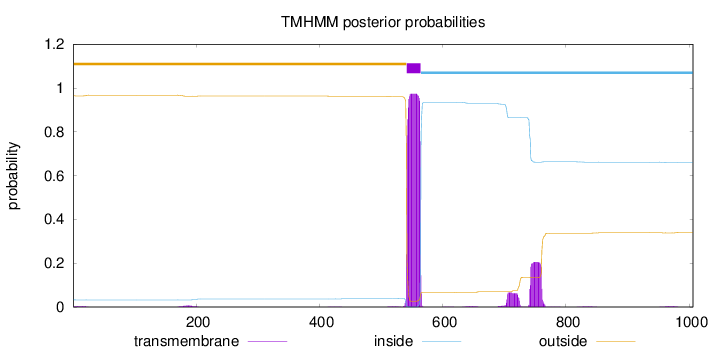
Length:
1006
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
27.98596
Exp number, first 60 AAs:
0.0714100000000001
Total prob of N-in:
0.03566
outside
1 - 541
TMhelix
542 - 564
inside
565 - 1006
Population Genetic Test Statistics
Pi
4.910585
Theta
4.002788
Tajima's D
0.626516
CLR
0.113201
CSRT
0.584370781460927
Interpretation
Uncertain
