Gene
KWMTBOMO00019 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002077
Annotation
PREDICTED:_tan_protein_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.47
Sequence
CDS
ATGAAACTATTGAATTACGGCGGAGTTGTTGCTATATTTTTAAGTTTATTCGGTACGGCAGTATGTGAGATACAACTCGGGAGACGCCATGCGGTCCCTGTTATATACGTAAGAGGCTCCCATTATGAAGTCGGCTTCGATGTGGGTCGCACATTCTCCTCCATCATCAAGAGCTTCATATCAAATTACGCTAATCTACGTGACTTTGAGCGCGAATACAAAACTGATACTGGCCGGAATGCGTACGATAAGACCCTGGCTAACATGGAGAAGCGATTCCCTTACTACGTTAAGGAGATCAGGGGTGTTGCTGATGGAGCTAACGTTCCTTTCTATCAGCTATTCCTCCTGCAAATGGATGACTTGATTGGAACCATCAACGACAACCACGTCCCACGGAACGACACCGGAGGCTGCAGTTCTGTCGCCTTCAAGAACCCACAACATACTATACTTGGACACACAGAGGATGCATTTACCGAAACCCTAAATCACTTCTACATTATGTCGGCGCACATCATTCCAACTCCCGAAGACAGGGAGCACGGGGCAGTAGAGGAACGGTTCTCGTCTTTATGCTACGCTGGTCACATGCCCGGGTACACCATGGGCTACAATGAAAACGGAATGGTGTTTTCCATCAACACGCTCAGCCCACTTCTGCTAAAGCCTGGAAACACTCCTCGCACATTTATAACCAGAAAGCTTCTGTCCGCCAAGAGTTTCCCGGATGCGGAACGGATCCTCCGTGACGAAGGCCTAGGAATCGGCAACGGTTTTTCGGTAAATATGATTTGGACAGACACCTGGGGGGATACCAAGCTTTACAATGTGGAAGTCGCCCCCGACCTCAAGGGAGACCGCTCGCTCGTCCACGTTCAGAAATACGACCAGGACACACTAGTACACTGTAATATTTACAACCGCATTAACGTGATTGAAGTGATTGGCGAAATAGTTGGCAGTAGCAAGTCCCGACTGAAGGCGATACACGCTCATGCTGCCCCCAAGACCAGAGGTGATATAGCTGAGATACTCTCTGATGTCACCGGCAAAGACTTCCAAGTGTTCTCGATGCAGAAGGACGCTATTATCAAGACCATCGCTGCTGGTATATTCGACGTAGAAAAAAGAACGTGGAGTATTTATATCGACAAACCCAGCATTTCGGAGCCCGTTGCAGTGCTGCCCATCAGATTCTCCGAGCTCGAGTCTAATAAATAA
Protein
MKLLNYGGVVAIFLSLFGTAVCEIQLGRRHAVPVIYVRGSHYEVGFDVGRTFSSIIKSFISNYANLRDFEREYKTDTGRNAYDKTLANMEKRFPYYVKEIRGVADGANVPFYQLFLLQMDDLIGTINDNHVPRNDTGGCSSVAFKNPQHTILGHTEDAFTETLNHFYIMSAHIIPTPEDREHGAVEERFSSLCYAGHMPGYTMGYNENGMVFSINTLSPLLLKPGNTPRTFITRKLLSAKSFPDAERILRDEGLGIGNGFSVNMIWTDTWGDTKLYNVEVAPDLKGDRSLVHVQKYDQDTLVHCNIYNRINVIEVIGEIVGSSKSRLKAIHAHAAPKTRGDIAEILSDVTGKDFQVFSMQKDAIIKTIAAGIFDVEKRTWSIYIDKPSISEPVAVLPIRFSELESNK
Summary
Uniprot
D4AH61
H9IXU4
F2VLM1
A0A0G3VJK4
A0A212EIC7
S4PX07
+ More
A0A2A4JK44 A0A2H1WTZ8 D5MRS0 D5MRR8 A0A194QZD7 D4AH58 A0A194PT80 D5LPT8 T1TE52 A0A1W4X4D5 K7J8V8 A0A232FFP0 A0A026VVP6 A0A290GLG3 D6WGK9 E2AV98 A0A0L7QSC8 U5ECS2 A0A336LZZ7 A0A0C9QJL3 A0A1J1HHG7 A0A2J7PUX3 A0A2J7PUW2 A0A088A3P1 A0A2A3E3A6 Q17I81 Q17I82 A0A1Q3FGX4 A0A154NYP4 A0A182MCH8 A0A023ESC5 A0A182R213 B0WJ49 A0A182VVJ5 A0A182KHA7 A0A182PNT7 A0A195EH99 A0A0N0BJ17 A0A158NHV9 A0A195AWZ9 A0A151WQV8 E2C8M5 A0A195FEL2 W5JJP1 N6T742 A0A195BY47 A0A2M4A0S4 A0A1B6DIH8 A0A182X2E3 E9IVF3 A0A2M4BR05 A0A182VFM7 A0A182LCX2 Q7QEK0 F4X3C1 A0A182FMK2 A0A1B6CEX5 A0A1Y1LDK7 A0A2M3Z741 A0A1B6LMT1 A0A0K8VPB4 A0A1L8DCS4 T1PEV9 A0A182N122 A0A182QY38 A0A034VLJ8 A0A034VIU1 A0A0A1WLD6 A0A1I8NCT6 W8B3Y4 A0A182HIU1 A0A182XYE7 A0A1B0CNI7 A0A182IPH6 A0A0L0CK10 A0A1L8EHR9 A0A1L8EHN7 A0A084WF21 A0A1J1IM57 E0VXG3 A0A182G696 C9E3G0 C9E3G6 C9E3F8 B4MET1 A0A1W4UQU8 C9E3F6 B4JJS6 C9E3G8 C9E3H0 C9E3G1 C9E3F7 Q9W369 C9E3G4
A0A2A4JK44 A0A2H1WTZ8 D5MRS0 D5MRR8 A0A194QZD7 D4AH58 A0A194PT80 D5LPT8 T1TE52 A0A1W4X4D5 K7J8V8 A0A232FFP0 A0A026VVP6 A0A290GLG3 D6WGK9 E2AV98 A0A0L7QSC8 U5ECS2 A0A336LZZ7 A0A0C9QJL3 A0A1J1HHG7 A0A2J7PUX3 A0A2J7PUW2 A0A088A3P1 A0A2A3E3A6 Q17I81 Q17I82 A0A1Q3FGX4 A0A154NYP4 A0A182MCH8 A0A023ESC5 A0A182R213 B0WJ49 A0A182VVJ5 A0A182KHA7 A0A182PNT7 A0A195EH99 A0A0N0BJ17 A0A158NHV9 A0A195AWZ9 A0A151WQV8 E2C8M5 A0A195FEL2 W5JJP1 N6T742 A0A195BY47 A0A2M4A0S4 A0A1B6DIH8 A0A182X2E3 E9IVF3 A0A2M4BR05 A0A182VFM7 A0A182LCX2 Q7QEK0 F4X3C1 A0A182FMK2 A0A1B6CEX5 A0A1Y1LDK7 A0A2M3Z741 A0A1B6LMT1 A0A0K8VPB4 A0A1L8DCS4 T1PEV9 A0A182N122 A0A182QY38 A0A034VLJ8 A0A034VIU1 A0A0A1WLD6 A0A1I8NCT6 W8B3Y4 A0A182HIU1 A0A182XYE7 A0A1B0CNI7 A0A182IPH6 A0A0L0CK10 A0A1L8EHR9 A0A1L8EHN7 A0A084WF21 A0A1J1IM57 E0VXG3 A0A182G696 C9E3G0 C9E3G6 C9E3F8 B4MET1 A0A1W4UQU8 C9E3F6 B4JJS6 C9E3G8 C9E3H0 C9E3G1 C9E3F7 Q9W369 C9E3G4
Pubmed
19121390
22118469
23622113
26354079
20075255
28648823
+ More
24508170 18362917 19820115 20798317 17510324 24945155 21347285 20920257 23761445 23537049 21282665 20966253 12364791 14747013 17210077 21719571 28004739 25348373 25830018 25315136 24495485 25244985 26108605 24438588 20566863 26483478 19900891 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21571070 26109357 26109356
24508170 18362917 19820115 20798317 17510324 24945155 21347285 20920257 23761445 23537049 21282665 20966253 12364791 14747013 17210077 21719571 28004739 25348373 25830018 25315136 24495485 25244985 26108605 24438588 20566863 26483478 19900891 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21571070 26109357 26109356
EMBL
AB499125
BAI87831.1
BABH01015619
BABH01015620
GU386341
ADU32897.1
+ More
KP657627 AKL78852.1 AGBW02014676 OWR41220.1 GAIX01005868 JAA86692.1 NWSH01001255 PCG71964.1 ODYU01011053 SOQ56545.1 AB531134 BAJ07601.1 AB531132 BAJ07599.1 KQ460953 KPJ10340.1 AB499122 BAI87828.1 KQ459599 KPI94340.1 GU953228 ADF43212.1 KC996755 AGT62465.1 AAZX01000808 NNAY01000336 OXU29117.1 KK107770 EZA47863.1 KY221866 ATB56364.1 KQ971321 EFA00578.1 GL443059 EFN62652.1 KQ414758 KOC61454.1 GANO01004998 JAB54873.1 UFQS01000373 UFQT01000373 SSX03289.1 SSX23656.1 GBYB01000747 JAG70514.1 CVRI01000004 CRK87323.1 NEVH01021191 PNF20124.1 PNF20122.1 KZ288403 PBC26195.1 CH477242 EAT46346.1 EAT46345.1 GFDL01008244 JAV26801.1 KQ434783 KZC04743.1 AXCM01015971 GAPW01001947 JAC11651.1 DS231955 EDS28928.1 KQ978957 KYN27247.1 KQ435724 KOX78492.1 ADTU01016076 ADTU01016077 ADTU01016078 KQ976725 KYM76577.1 KQ982813 KYQ50292.1 GL453666 EFN75755.1 KQ981636 KYN38816.1 ADMH02000949 ETN64592.1 APGK01041353 KB740993 ENN76034.1 KQ978501 KYM93584.1 GGFK01000999 MBW34320.1 GEDC01019220 GEDC01013169 GEDC01011804 GEDC01005456 JAS18078.1 JAS24129.1 JAS25494.1 JAS31842.1 GL766213 EFZ15450.1 GGFJ01006311 MBW55452.1 AAAB01008846 EAA06499.3 GL888609 EGI59102.1 GEDC01025339 GEDC01013267 JAS11959.1 JAS24031.1 GEZM01062088 JAV70045.1 GGFM01003570 MBW24321.1 GEBQ01015173 JAT24804.1 GDHF01011894 JAI40420.1 GFDF01009917 JAV04167.1 KA647276 AFP61905.1 AXCN02001385 GAKP01016529 JAC42423.1 GAKP01016528 JAC42424.1 GBXI01014992 JAC99299.1 GAMC01010705 JAB95850.1 APCN01004096 AJWK01020382 AJWK01020383 AJWK01020384 AJWK01020385 AJWK01020386 AXCP01008174 JRES01000302 KNC32552.1 GFDG01000633 JAV18166.1 GFDG01000634 JAV18165.1 ATLV01023287 KE525342 KFB48815.1 CVRI01000051 CRK99553.1 DS235832 EEB18069.1 JXUM01044516 JXUM01044517 KQ561404 KXJ78689.1 GQ457340 ACV86748.1 GQ457346 ACV86754.1 GQ457338 GQ457339 GQ457347 ACV86746.1 ACV86747.1 ACV86755.1 CH940664 EDW63056.1 GQ457336 ACV86744.1 CH916370 EDV99828.1 GQ457348 ACV86756.1 GQ457350 ACV86758.1 GQ457341 GQ457342 GQ457343 GQ457349 ACV86749.1 ACV86750.1 ACV86751.1 ACV86757.1 GQ457337 GQ457351 GQ457353 ACV86745.1 ACV86759.1 ACV86761.1 AE014298 AY128492 GU144522 AAF46466.1 AAM75085.1 ACZ36950.1 AHN59513.1 GQ457344 ACV86752.1
KP657627 AKL78852.1 AGBW02014676 OWR41220.1 GAIX01005868 JAA86692.1 NWSH01001255 PCG71964.1 ODYU01011053 SOQ56545.1 AB531134 BAJ07601.1 AB531132 BAJ07599.1 KQ460953 KPJ10340.1 AB499122 BAI87828.1 KQ459599 KPI94340.1 GU953228 ADF43212.1 KC996755 AGT62465.1 AAZX01000808 NNAY01000336 OXU29117.1 KK107770 EZA47863.1 KY221866 ATB56364.1 KQ971321 EFA00578.1 GL443059 EFN62652.1 KQ414758 KOC61454.1 GANO01004998 JAB54873.1 UFQS01000373 UFQT01000373 SSX03289.1 SSX23656.1 GBYB01000747 JAG70514.1 CVRI01000004 CRK87323.1 NEVH01021191 PNF20124.1 PNF20122.1 KZ288403 PBC26195.1 CH477242 EAT46346.1 EAT46345.1 GFDL01008244 JAV26801.1 KQ434783 KZC04743.1 AXCM01015971 GAPW01001947 JAC11651.1 DS231955 EDS28928.1 KQ978957 KYN27247.1 KQ435724 KOX78492.1 ADTU01016076 ADTU01016077 ADTU01016078 KQ976725 KYM76577.1 KQ982813 KYQ50292.1 GL453666 EFN75755.1 KQ981636 KYN38816.1 ADMH02000949 ETN64592.1 APGK01041353 KB740993 ENN76034.1 KQ978501 KYM93584.1 GGFK01000999 MBW34320.1 GEDC01019220 GEDC01013169 GEDC01011804 GEDC01005456 JAS18078.1 JAS24129.1 JAS25494.1 JAS31842.1 GL766213 EFZ15450.1 GGFJ01006311 MBW55452.1 AAAB01008846 EAA06499.3 GL888609 EGI59102.1 GEDC01025339 GEDC01013267 JAS11959.1 JAS24031.1 GEZM01062088 JAV70045.1 GGFM01003570 MBW24321.1 GEBQ01015173 JAT24804.1 GDHF01011894 JAI40420.1 GFDF01009917 JAV04167.1 KA647276 AFP61905.1 AXCN02001385 GAKP01016529 JAC42423.1 GAKP01016528 JAC42424.1 GBXI01014992 JAC99299.1 GAMC01010705 JAB95850.1 APCN01004096 AJWK01020382 AJWK01020383 AJWK01020384 AJWK01020385 AJWK01020386 AXCP01008174 JRES01000302 KNC32552.1 GFDG01000633 JAV18166.1 GFDG01000634 JAV18165.1 ATLV01023287 KE525342 KFB48815.1 CVRI01000051 CRK99553.1 DS235832 EEB18069.1 JXUM01044516 JXUM01044517 KQ561404 KXJ78689.1 GQ457340 ACV86748.1 GQ457346 ACV86754.1 GQ457338 GQ457339 GQ457347 ACV86746.1 ACV86747.1 ACV86755.1 CH940664 EDW63056.1 GQ457336 ACV86744.1 CH916370 EDV99828.1 GQ457348 ACV86756.1 GQ457350 ACV86758.1 GQ457341 GQ457342 GQ457343 GQ457349 ACV86749.1 ACV86750.1 ACV86751.1 ACV86757.1 GQ457337 GQ457351 GQ457353 ACV86745.1 ACV86759.1 ACV86761.1 AE014298 AY128492 GU144522 AAF46466.1 AAM75085.1 ACZ36950.1 AHN59513.1 GQ457344 ACV86752.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000053268
UP000192223
+ More
UP000002358 UP000215335 UP000053097 UP000007266 UP000000311 UP000053825 UP000183832 UP000235965 UP000005203 UP000242457 UP000008820 UP000076502 UP000075883 UP000075900 UP000002320 UP000075920 UP000075881 UP000075885 UP000078492 UP000053105 UP000005205 UP000078540 UP000075809 UP000008237 UP000078541 UP000000673 UP000019118 UP000078542 UP000076407 UP000075903 UP000075882 UP000007062 UP000007755 UP000069272 UP000075884 UP000075886 UP000095301 UP000075840 UP000076408 UP000092461 UP000075880 UP000037069 UP000030765 UP000009046 UP000069940 UP000249989 UP000008792 UP000192221 UP000001070 UP000000803
UP000002358 UP000215335 UP000053097 UP000007266 UP000000311 UP000053825 UP000183832 UP000235965 UP000005203 UP000242457 UP000008820 UP000076502 UP000075883 UP000075900 UP000002320 UP000075920 UP000075881 UP000075885 UP000078492 UP000053105 UP000005205 UP000078540 UP000075809 UP000008237 UP000078541 UP000000673 UP000019118 UP000078542 UP000076407 UP000075903 UP000075882 UP000007062 UP000007755 UP000069272 UP000075884 UP000075886 UP000095301 UP000075840 UP000076408 UP000092461 UP000075880 UP000037069 UP000030765 UP000009046 UP000069940 UP000249989 UP000008792 UP000192221 UP000001070 UP000000803
Pfam
PF03417 AAT
Interpro
IPR005079
Peptidase_C45
ProteinModelPortal
D4AH61
H9IXU4
F2VLM1
A0A0G3VJK4
A0A212EIC7
S4PX07
+ More
A0A2A4JK44 A0A2H1WTZ8 D5MRS0 D5MRR8 A0A194QZD7 D4AH58 A0A194PT80 D5LPT8 T1TE52 A0A1W4X4D5 K7J8V8 A0A232FFP0 A0A026VVP6 A0A290GLG3 D6WGK9 E2AV98 A0A0L7QSC8 U5ECS2 A0A336LZZ7 A0A0C9QJL3 A0A1J1HHG7 A0A2J7PUX3 A0A2J7PUW2 A0A088A3P1 A0A2A3E3A6 Q17I81 Q17I82 A0A1Q3FGX4 A0A154NYP4 A0A182MCH8 A0A023ESC5 A0A182R213 B0WJ49 A0A182VVJ5 A0A182KHA7 A0A182PNT7 A0A195EH99 A0A0N0BJ17 A0A158NHV9 A0A195AWZ9 A0A151WQV8 E2C8M5 A0A195FEL2 W5JJP1 N6T742 A0A195BY47 A0A2M4A0S4 A0A1B6DIH8 A0A182X2E3 E9IVF3 A0A2M4BR05 A0A182VFM7 A0A182LCX2 Q7QEK0 F4X3C1 A0A182FMK2 A0A1B6CEX5 A0A1Y1LDK7 A0A2M3Z741 A0A1B6LMT1 A0A0K8VPB4 A0A1L8DCS4 T1PEV9 A0A182N122 A0A182QY38 A0A034VLJ8 A0A034VIU1 A0A0A1WLD6 A0A1I8NCT6 W8B3Y4 A0A182HIU1 A0A182XYE7 A0A1B0CNI7 A0A182IPH6 A0A0L0CK10 A0A1L8EHR9 A0A1L8EHN7 A0A084WF21 A0A1J1IM57 E0VXG3 A0A182G696 C9E3G0 C9E3G6 C9E3F8 B4MET1 A0A1W4UQU8 C9E3F6 B4JJS6 C9E3G8 C9E3H0 C9E3G1 C9E3F7 Q9W369 C9E3G4
A0A2A4JK44 A0A2H1WTZ8 D5MRS0 D5MRR8 A0A194QZD7 D4AH58 A0A194PT80 D5LPT8 T1TE52 A0A1W4X4D5 K7J8V8 A0A232FFP0 A0A026VVP6 A0A290GLG3 D6WGK9 E2AV98 A0A0L7QSC8 U5ECS2 A0A336LZZ7 A0A0C9QJL3 A0A1J1HHG7 A0A2J7PUX3 A0A2J7PUW2 A0A088A3P1 A0A2A3E3A6 Q17I81 Q17I82 A0A1Q3FGX4 A0A154NYP4 A0A182MCH8 A0A023ESC5 A0A182R213 B0WJ49 A0A182VVJ5 A0A182KHA7 A0A182PNT7 A0A195EH99 A0A0N0BJ17 A0A158NHV9 A0A195AWZ9 A0A151WQV8 E2C8M5 A0A195FEL2 W5JJP1 N6T742 A0A195BY47 A0A2M4A0S4 A0A1B6DIH8 A0A182X2E3 E9IVF3 A0A2M4BR05 A0A182VFM7 A0A182LCX2 Q7QEK0 F4X3C1 A0A182FMK2 A0A1B6CEX5 A0A1Y1LDK7 A0A2M3Z741 A0A1B6LMT1 A0A0K8VPB4 A0A1L8DCS4 T1PEV9 A0A182N122 A0A182QY38 A0A034VLJ8 A0A034VIU1 A0A0A1WLD6 A0A1I8NCT6 W8B3Y4 A0A182HIU1 A0A182XYE7 A0A1B0CNI7 A0A182IPH6 A0A0L0CK10 A0A1L8EHR9 A0A1L8EHN7 A0A084WF21 A0A1J1IM57 E0VXG3 A0A182G696 C9E3G0 C9E3G6 C9E3F8 B4MET1 A0A1W4UQU8 C9E3F6 B4JJS6 C9E3G8 C9E3H0 C9E3G1 C9E3F7 Q9W369 C9E3G4
PDB
2X1E
E-value=0.000185049,
Score=106
Ontologies
GO
Topology
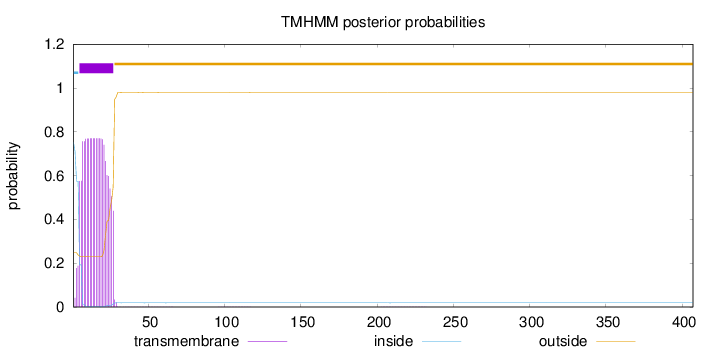
SignalP
Position: 1 - 22,
Likelihood: 0.936115

Length:
407
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
16.4904299999999
Exp number, first 60 AAs:
16.46908
Total prob of N-in:
0.75212
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 407
Population Genetic Test Statistics
Pi
28.830895
Theta
25.222795
Tajima's D
0.195858
CLR
0.040738
CSRT
0.431328433578321
Interpretation
Uncertain
