Gene
KWMTBOMO00014
Pre Gene Modal
BGIBMGA012989
Annotation
Yokozuna_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.669
Sequence
CDS
ATGGCGTCAACAGTTCCAGTAACAAACGCCATAAATTATTACCCACAACTTAACGTGGAAAATTACAATAGCTGGAAGTTTCGACTGAGATGTCTATTAGACGAAAAATCTTTGTTAGAGGTAGCGTCAAGTGGCGTTATAGAATTAAAACAAAAAACTGCTGATGCGGCTGCAAAAGCCTTAATAGTAAAATGTGTACCCGATAAGTACTTGGACATCATAAAAGATGCAACAACTAGCTACACAATGTTAGCCAGTCTGGATAAAGTTTTTGAGAGAAAAGGTATTTTCACTAAATTACACTTGCGAAGAAAGTTATTGTCTCTTAAGCTGAGGGATGATAAATTGGAAGACTATTTCATGAGATTTGAAAGTCTGATTAGAGAAATAGAAAATATTGATAAAAAGATGGATGAAGAAGATAAAATATGTTATTTGCTGACGGGTATGTCGGAAAAGTACAACACAGTGATAACTGCTATTGAAACAATGTCTGCAGATAAAATTGACATGGAGTTTGTCAAGGCTCGGTTATTGGACGAAGAACTCAAACTGAATAATAAAGGTGAAGAGTATAATGATACAAAGAATGATGTAAGTTTTACTGCCTCAAGTGTAGAATGTTATCGTTGCCACAAAAAGGGCCATTTTGCACGTGAATGTAGAAGCAGAGGTTGTGGCTATTCAAGGAGAAACATTAATAATAAAGGTATATGTGGTAAAAACAGAACTGAAGAACAAAATAATATGACAGAAAGCATCCCAGAGAATAAATATAATTTTCTAGCAATAACGGAGTGTAATCTAAGTAAAGAAGACAAATTTATAGAATTTATAATAGATTCAGGAGCTACAGAAAATCTTATCAAGGAAGAATTGGAGAAATACATGTATGAAATTAAGAAACTACCAAATAAAATAAAAATTAAGATTGCAAATGGAGAAATATTAGAGGCTGACAAGAAAGGAAAGATAACCATAAGAGATAAAAATAATAATACTATAAATATAGAAGCGTTAATGGTCCCAGGGATTTATCATAATATAATATCTGTAAGAAACTTAAATATGAAAGGAAATAAAGTTGTTTTTACAAAATTTGGAGCACAAATAATAAATGGAAGAAAAGTCTTAAAATGTTCAGATAGAAATGGTTTATATATAGTACAAGGCGAAATAATACTAATAGAAGAAGCGAACTCAAGTGTACAGCAAGACAAAAATATTTGGCACCAACGTCTCGGACACTTAAATAGGAAATCATTAAAAATTATGGGTTTACCTGTGTCTGATGAAACATGTGGTCCTTGCATGAAGGGAAAATCTACTCGTTTACCATTCTATCCAGTTGACAGGCCAAGAAGTAGAAAAATAGGAGAATTATTGCATACAGATATTGGAGGTCCTGTATCACCAATTTCTTATGATGAAAAACAATATTACCAGACCATAACTGATGATTTTTCTCATTTTGTGACTGTTTATCTATTAAAGAATAAATCTGAAGCCACAAGAAATTTAATAAATCATATAAAGGAGTTAGAAAGACAAAAGGAAACTAAAGTTAAACGCATAAGATGTGATAATGGTGGAGAGTTCACCGCTGGCTGTTTGAAGCAATTTTGTATGGAAAATGGAATTATAATAGAGTATAGTCAACCTTACACACCACAACAAAATGGTGTGGCAGAGAGGCTAAACAGAACATTACTAGATAAGTCAAGAACTTTATTAAGTGAAACTGGATTACCTAAATATTTGTGGTCTGAGGCTATAATGTGTGCAGCATATCAATTGAATCGATGCCCATCAAGATCTATAAAGTATAAGATACCAGCTGAAATATACTACGGACTAGAAAATGTTAACTTGAACAAGTTAAAGGTATTTGGTTCTAAAGCATGGTATACGTGTGTCCCACAGAATAAGAAATTAGATAATAGAGCTGAAGAAGCCATTATGGTTGGCTATGGCAAACAAGGTTATAGATTATGGAATCCAAAAACAACAGAAATAATATTCTGCAGAGATGTCAAGTTTGATGAAAAGTACTATGAACATAAAAACAGTATTAAAAATAACAACTATATTTATTTACCAGAAGAGGACACACAAAATGAAGTTATGTGCAATGATAAAAATAAAATTGAGGAAGAAAAAGTAATACAGAAAAATGAAGAGACAGACACAAAAAGAACTAGATCAGGAAGGATAATAAAGCAACCTGATTATCTAAAGAATTTTGGGACGTACACAGCATATTGTTACTTAGTGTGTAATGACCCAGAAAGCTATGAAGAAGCGATAAAAGAAGTTTCATGGAAAGAAGCGATACATAAGGAATTAGCAGCACTTGAGAACTTACAAAGCTGGGAAGAGACTAAACTACCGAAAGGAAAATTAGCTATAGACACTAAGTGGGTTTTTAGAACGAAAGACGATGGCACTAAGAAAGCCAGATTAGTCGCTCGAGGTTTTCAAATAAAAAATTATGAGAATGAAATATTCTACAGTCCTGTAGTAAGAATGTCTACGATAAGAGCATTTTTTGTAAAAGCTATCCAAGAAGGATACAAATTAAGGCAATTGGATATTCCTACAGCCTTTCTCAATGGATTTCTTGATAGTGAAGTTTATATAAAAATACCCAAAGGCAAAGAAAACAAAGATGGAATAGTTTTGAGGTTAAAGAGAGCATTGTATGGATTAAAAGAAAGCCCTAAATGCTGGAACATGAGATTTGATGAATTTGCAAGAAAGAATGGATTGATACAGTCATTACATGATTGTTGTCTATACACAGATAAAAATATATGGCTTATAATATTTGTAGATGATATTTTATTAATTGGAATGGAAAAGGATACAGAAAAAATTATTAATAAATTGAAGCAAGAATTTAAAGCTAAAGATTTAGGACAGATTGAGAAATATCTTGGTATGGAAATTAAACAGGAAGAAAGTAAATTAACGATTAAACAAGAAAAGATAATTGAGAAGATACTAGAAAAATTTGGAATGTCAGAATGTAAAGGTATAGGAACACCTATGGAGGTAAATTTTCAAATAAACAAAAATGAGAAGATTATTGATGTACCATATCGACAACTCATAGGAAGCATAATGTATTTGGCTACTATGTCCAGACCAGATCTGCAGTTTGCTGTATCATATCTTAGCAGATACCTAGACTGTCCTACTCAACAGACTTGGAAAGCTGGAAAGAGAATTCTGCAATACCTAAAAGCTACAAAAGACAAAATGTTAGTTTTTCAAAAAAGGAATAATCTAGATGTGGATTTATGTGCATATTCAGACTCTGATTGGGCAGGTGATATTGTTGATCGCAAAAGTGTCAGTGGGGGTATCATAATGTGTAGTGATAATCCTGTATTTTGGTACTCTAGAAAACAAAGTTGTGTTGCATTATCGACAGCAGAATCAGAGTACATTGCTAGTGCTTGTTGTGCTCAAGAGTTAGTGTATTTGAAAGGTGTACTGTGTGATTTTGGATATGAATGTAAATTGACTTTATTTATGGATAATAAGGGAGCAATATGTTTAGCGAAGTCAAATGAAAACTCTAAAAGGAGTAAGCATATTGATATAAGGATGCACTATATTAAGGACTTGGTGAGGAATAATGTATTAGATATTGAATATGTGTCATCTGATAAAAATATTGCTGATATTTGTACAAAATCTTTATGCAAAGAAAAGTTTGTATTTTTTAGAAGTTTGTTACTAAAGTAA
Protein
MASTVPVTNAINYYPQLNVENYNSWKFRLRCLLDEKSLLEVASSGVIELKQKTADAAAKALIVKCVPDKYLDIIKDATTSYTMLASLDKVFERKGIFTKLHLRRKLLSLKLRDDKLEDYFMRFESLIREIENIDKKMDEEDKICYLLTGMSEKYNTVITAIETMSADKIDMEFVKARLLDEELKLNNKGEEYNDTKNDVSFTASSVECYRCHKKGHFARECRSRGCGYSRRNINNKGICGKNRTEEQNNMTESIPENKYNFLAITECNLSKEDKFIEFIIDSGATENLIKEELEKYMYEIKKLPNKIKIKIANGEILEADKKGKITIRDKNNNTINIEALMVPGIYHNIISVRNLNMKGNKVVFTKFGAQIINGRKVLKCSDRNGLYIVQGEIILIEEANSSVQQDKNIWHQRLGHLNRKSLKIMGLPVSDETCGPCMKGKSTRLPFYPVDRPRSRKIGELLHTDIGGPVSPISYDEKQYYQTITDDFSHFVTVYLLKNKSEATRNLINHIKELERQKETKVKRIRCDNGGEFTAGCLKQFCMENGIIIEYSQPYTPQQNGVAERLNRTLLDKSRTLLSETGLPKYLWSEAIMCAAYQLNRCPSRSIKYKIPAEIYYGLENVNLNKLKVFGSKAWYTCVPQNKKLDNRAEEAIMVGYGKQGYRLWNPKTTEIIFCRDVKFDEKYYEHKNSIKNNNYIYLPEEDTQNEVMCNDKNKIEEEKVIQKNEETDTKRTRSGRIIKQPDYLKNFGTYTAYCYLVCNDPESYEEAIKEVSWKEAIHKELAALENLQSWEETKLPKGKLAIDTKWVFRTKDDGTKKARLVARGFQIKNYENEIFYSPVVRMSTIRAFFVKAIQEGYKLRQLDIPTAFLNGFLDSEVYIKIPKGKENKDGIVLRLKRALYGLKESPKCWNMRFDEFARKNGLIQSLHDCCLYTDKNIWLIIFVDDILLIGMEKDTEKIINKLKQEFKAKDLGQIEKYLGMEIKQEESKLTIKQEKIIEKILEKFGMSECKGIGTPMEVNFQINKNEKIIDVPYRQLIGSIMYLATMSRPDLQFAVSYLSRYLDCPTQQTWKAGKRILQYLKATKDKMLVFQKRNNLDVDLCAYSDSDWAGDIVDRKSVSGGIIMCSDNPVFWYSRKQSCVALSTAESEYIASACCAQELVYLKGVLCDFGYECKLTLFMDNKGAICLAKSNENSKRSKHIDIRMHYIKDLVRNNVLDIEYVSSDKNIADICTKSLCKEKFVFFRSLLLK
Summary
Uniprot
O96968
A0A146L6N5
A0A0A9XG27
A0A146KTS1
A0A1B6DFK3
A0A0A9Z0I6
+ More
A0A0A9WLY1 A0A1Y1L115 A0A1Y1MQF0 A0A224XA26 A0A182GA29 J9KJ72 A0A0A9ZBW6 A0A023F102 A0A0T6AT79 A0A182H4X3 A0A1W7R6I5 A0A068B703 A0A182G1G7 A0A1Y1MJE9 A0A1B6C2D3 A0A151RCT9 A0A151TPU3 A0A151RPT4 A0A182GGE6 Q967L5 A0A0J7K6U3 A0A2N9IUL9 A0A1W7R6A3 A0A151TGP0 A0A0A9W6B0 A0A2N9JB15 A0A151TNK0 A0A146LT61 A0A1Y1LCN3 B8YLY6 Q9C739 Q9LH44 A0A2N9GX95 A0A2N9GHK9 A0A0N1ICY1 A0A151QNH5 A0A151RJN4 A0A151TMN9 A0A224XJ98 B8YLY4 A0A151SDY6 B8YLY3 A0A059QBK0 Q9SXB2 A0A1Y1M2G6 A0A0A9XEU3 A0A151R6U8 Q9C536 A0A194RJV9 A0A0J7K6W1 B8YLY5 Q9M2D1 A0A0V0RWJ3 A0A2Z6NRJ0 A0A2N9GHB5 A0A2N9F1W0 A0A151RAL1 A0A194RU38 A0A034VP82 A0A087SVE4 A0A151R2G6 A0A2A4J959 A0A131XNF9 A0A2R6RYD4 A0A1Y1MYL8 A0A151QZS0 A0A0P6K133 A0A0P6J144 A0A151TKT1 A0A061F3M7 A0A1Y1MYL6 A0A151T469 A0A0V0TUP0 A0A2K3N1R3 A0A2N9ESI4 A0A2N9J3Y8 E5SJ15 A0A151RBY1 E5SZD6 Q6L3N8 A0A1W7R685 A0A2N9HHD8 A0A151S6I6 E5SB91 A0A2K3MZ63 A0A2N9I1L5 A0A2N9FNY4 A0A151TIH6 A0A0V1APS7 A0A151SHZ9
A0A0A9WLY1 A0A1Y1L115 A0A1Y1MQF0 A0A224XA26 A0A182GA29 J9KJ72 A0A0A9ZBW6 A0A023F102 A0A0T6AT79 A0A182H4X3 A0A1W7R6I5 A0A068B703 A0A182G1G7 A0A1Y1MJE9 A0A1B6C2D3 A0A151RCT9 A0A151TPU3 A0A151RPT4 A0A182GGE6 Q967L5 A0A0J7K6U3 A0A2N9IUL9 A0A1W7R6A3 A0A151TGP0 A0A0A9W6B0 A0A2N9JB15 A0A151TNK0 A0A146LT61 A0A1Y1LCN3 B8YLY6 Q9C739 Q9LH44 A0A2N9GX95 A0A2N9GHK9 A0A0N1ICY1 A0A151QNH5 A0A151RJN4 A0A151TMN9 A0A224XJ98 B8YLY4 A0A151SDY6 B8YLY3 A0A059QBK0 Q9SXB2 A0A1Y1M2G6 A0A0A9XEU3 A0A151R6U8 Q9C536 A0A194RJV9 A0A0J7K6W1 B8YLY5 Q9M2D1 A0A0V0RWJ3 A0A2Z6NRJ0 A0A2N9GHB5 A0A2N9F1W0 A0A151RAL1 A0A194RU38 A0A034VP82 A0A087SVE4 A0A151R2G6 A0A2A4J959 A0A131XNF9 A0A2R6RYD4 A0A1Y1MYL8 A0A151QZS0 A0A0P6K133 A0A0P6J144 A0A151TKT1 A0A061F3M7 A0A1Y1MYL6 A0A151T469 A0A0V0TUP0 A0A2K3N1R3 A0A2N9ESI4 A0A2N9J3Y8 E5SJ15 A0A151RBY1 E5SZD6 Q6L3N8 A0A1W7R685 A0A2N9HHD8 A0A151S6I6 E5SB91 A0A2K3MZ63 A0A2N9I1L5 A0A2N9FNY4 A0A151TIH6 A0A0V1APS7 A0A151SHZ9
Pubmed
EMBL
AB014676
BAA74713.1
GDHC01015802
JAQ02827.1
GBHO01027564
JAG16040.1
+ More
GDHC01019310 JAP99318.1 GEDC01012841 GEDC01000503 JAS24457.1 JAS36795.1 GBHO01004832 JAG38772.1 GBHO01035173 JAG08431.1 GEZM01072005 JAV65635.1 GEZM01028916 JAV86196.1 GFTR01008662 JAW07764.1 JXUM01155348 KQ572178 KXJ68066.1 ABLF02041793 GBHO01002761 JAG40843.1 GBBI01004033 JAC14679.1 LJIG01022881 KRT78250.1 JXUM01025857 KQ560736 KXJ81076.1 GEHC01000878 JAV46767.1 KF740825 AIC77183.1 JXUM01136904 KQ568499 KXJ68982.1 GEZM01032204 JAV84770.1 GEDC01029908 JAS07390.1 KQ483843 KYP40337.1 CM003606 KYP69041.1 KQ483623 KYP44533.1 JXUM01011189 KQ560325 KXJ83029.1 AY009101 AAK12626.1 LBMM01012492 KMQ86193.1 OIVN01006211 SPD27863.1 GEHC01000959 JAV46686.1 CM003608 KYP66220.1 GBHO01040663 JAG02941.1 OIVN01006466 SPD33659.1 KYP68607.1 GDHC01007678 JAQ10951.1 GEZM01063091 JAV69316.1 FJ544856 ACL97386.1 AC073555 AAG60117.1 AP002064 BAB01972.1 OIVN01002473 SPD03944.1 OIVN01001931 SPC99063.1 KQ460882 KPJ11482.1 KQ485638 KYP31826.1 KQ483702 KYP42748.1 KYP68287.1 GFTR01008327 JAW08099.1 FJ544853 ACL97384.1 KQ483417 KYP53032.1 FJ544851 ACL97383.1 KF303287 AGW47867.1 AC007259 AAD50001.1 GEZM01044912 JAV78046.1 GBHO01024362 JAG19242.1 KQ484038 KYP38095.1 AC079604 AC079131 AAG50698.1 AAG50765.1 KQ460297 KPJ16236.1 LBMM01012645 KMQ86082.1 FJ544854 ACL97385.1 AL137898 CAB71063.1 JYDL01000068 KRX18751.1 DF973742 GAU39052.1 OIVN01001906 SPC98838.1 OIVN01000486 SPC81000.1 KQ483890 KYP39660.1 KQ459875 KPJ19636.1 GAKP01015342 JAC43610.1 KK112142 KFM56833.1 KQ484186 KYP36645.1 NWSH01002292 PCG68657.1 PCG68658.1 GEFH01000951 JAP67630.1 NKQK01000002 PSS35035.1 GEZM01017328 JAV90763.1 KQ484324 KYP35753.1 GDUN01000215 JAN95704.1 GDUN01000217 JAN95702.1 CM003607 KYP67664.1 CM001883 EOY11267.1 GEZM01017329 JAV90762.1 CM003610 KYP61818.1 KYP63036.1 JYDJ01000136 KRX42725.1 ASHM01015075 PNX96991.1 OIVN01000279 SPC77544.1 OIVN01006353 SPD31284.1 KQ483853 KYP40164.1 AC149291 AAT38758.1 GEHC01000966 JAV46679.1 OIVN01003431 SPD11153.1 KQ483456 KYP50361.1 ASHM01014140 PNX96091.1 OIVN01004651 SPD18552.1 OIVN01001308 SPC92446.1 KYP66838.1 JYDH01000330 KRY26802.1 CM003613 KYP54398.1
GDHC01019310 JAP99318.1 GEDC01012841 GEDC01000503 JAS24457.1 JAS36795.1 GBHO01004832 JAG38772.1 GBHO01035173 JAG08431.1 GEZM01072005 JAV65635.1 GEZM01028916 JAV86196.1 GFTR01008662 JAW07764.1 JXUM01155348 KQ572178 KXJ68066.1 ABLF02041793 GBHO01002761 JAG40843.1 GBBI01004033 JAC14679.1 LJIG01022881 KRT78250.1 JXUM01025857 KQ560736 KXJ81076.1 GEHC01000878 JAV46767.1 KF740825 AIC77183.1 JXUM01136904 KQ568499 KXJ68982.1 GEZM01032204 JAV84770.1 GEDC01029908 JAS07390.1 KQ483843 KYP40337.1 CM003606 KYP69041.1 KQ483623 KYP44533.1 JXUM01011189 KQ560325 KXJ83029.1 AY009101 AAK12626.1 LBMM01012492 KMQ86193.1 OIVN01006211 SPD27863.1 GEHC01000959 JAV46686.1 CM003608 KYP66220.1 GBHO01040663 JAG02941.1 OIVN01006466 SPD33659.1 KYP68607.1 GDHC01007678 JAQ10951.1 GEZM01063091 JAV69316.1 FJ544856 ACL97386.1 AC073555 AAG60117.1 AP002064 BAB01972.1 OIVN01002473 SPD03944.1 OIVN01001931 SPC99063.1 KQ460882 KPJ11482.1 KQ485638 KYP31826.1 KQ483702 KYP42748.1 KYP68287.1 GFTR01008327 JAW08099.1 FJ544853 ACL97384.1 KQ483417 KYP53032.1 FJ544851 ACL97383.1 KF303287 AGW47867.1 AC007259 AAD50001.1 GEZM01044912 JAV78046.1 GBHO01024362 JAG19242.1 KQ484038 KYP38095.1 AC079604 AC079131 AAG50698.1 AAG50765.1 KQ460297 KPJ16236.1 LBMM01012645 KMQ86082.1 FJ544854 ACL97385.1 AL137898 CAB71063.1 JYDL01000068 KRX18751.1 DF973742 GAU39052.1 OIVN01001906 SPC98838.1 OIVN01000486 SPC81000.1 KQ483890 KYP39660.1 KQ459875 KPJ19636.1 GAKP01015342 JAC43610.1 KK112142 KFM56833.1 KQ484186 KYP36645.1 NWSH01002292 PCG68657.1 PCG68658.1 GEFH01000951 JAP67630.1 NKQK01000002 PSS35035.1 GEZM01017328 JAV90763.1 KQ484324 KYP35753.1 GDUN01000215 JAN95704.1 GDUN01000217 JAN95702.1 CM003607 KYP67664.1 CM001883 EOY11267.1 GEZM01017329 JAV90762.1 CM003610 KYP61818.1 KYP63036.1 JYDJ01000136 KRX42725.1 ASHM01015075 PNX96991.1 OIVN01000279 SPC77544.1 OIVN01006353 SPD31284.1 KQ483853 KYP40164.1 AC149291 AAT38758.1 GEHC01000966 JAV46679.1 OIVN01003431 SPD11153.1 KQ483456 KYP50361.1 ASHM01014140 PNX96091.1 OIVN01004651 SPD18552.1 OIVN01001308 SPC92446.1 KYP66838.1 JYDH01000330 KRY26802.1 CM003613 KYP54398.1
Proteomes
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
O96968
A0A146L6N5
A0A0A9XG27
A0A146KTS1
A0A1B6DFK3
A0A0A9Z0I6
+ More
A0A0A9WLY1 A0A1Y1L115 A0A1Y1MQF0 A0A224XA26 A0A182GA29 J9KJ72 A0A0A9ZBW6 A0A023F102 A0A0T6AT79 A0A182H4X3 A0A1W7R6I5 A0A068B703 A0A182G1G7 A0A1Y1MJE9 A0A1B6C2D3 A0A151RCT9 A0A151TPU3 A0A151RPT4 A0A182GGE6 Q967L5 A0A0J7K6U3 A0A2N9IUL9 A0A1W7R6A3 A0A151TGP0 A0A0A9W6B0 A0A2N9JB15 A0A151TNK0 A0A146LT61 A0A1Y1LCN3 B8YLY6 Q9C739 Q9LH44 A0A2N9GX95 A0A2N9GHK9 A0A0N1ICY1 A0A151QNH5 A0A151RJN4 A0A151TMN9 A0A224XJ98 B8YLY4 A0A151SDY6 B8YLY3 A0A059QBK0 Q9SXB2 A0A1Y1M2G6 A0A0A9XEU3 A0A151R6U8 Q9C536 A0A194RJV9 A0A0J7K6W1 B8YLY5 Q9M2D1 A0A0V0RWJ3 A0A2Z6NRJ0 A0A2N9GHB5 A0A2N9F1W0 A0A151RAL1 A0A194RU38 A0A034VP82 A0A087SVE4 A0A151R2G6 A0A2A4J959 A0A131XNF9 A0A2R6RYD4 A0A1Y1MYL8 A0A151QZS0 A0A0P6K133 A0A0P6J144 A0A151TKT1 A0A061F3M7 A0A1Y1MYL6 A0A151T469 A0A0V0TUP0 A0A2K3N1R3 A0A2N9ESI4 A0A2N9J3Y8 E5SJ15 A0A151RBY1 E5SZD6 Q6L3N8 A0A1W7R685 A0A2N9HHD8 A0A151S6I6 E5SB91 A0A2K3MZ63 A0A2N9I1L5 A0A2N9FNY4 A0A151TIH6 A0A0V1APS7 A0A151SHZ9
A0A0A9WLY1 A0A1Y1L115 A0A1Y1MQF0 A0A224XA26 A0A182GA29 J9KJ72 A0A0A9ZBW6 A0A023F102 A0A0T6AT79 A0A182H4X3 A0A1W7R6I5 A0A068B703 A0A182G1G7 A0A1Y1MJE9 A0A1B6C2D3 A0A151RCT9 A0A151TPU3 A0A151RPT4 A0A182GGE6 Q967L5 A0A0J7K6U3 A0A2N9IUL9 A0A1W7R6A3 A0A151TGP0 A0A0A9W6B0 A0A2N9JB15 A0A151TNK0 A0A146LT61 A0A1Y1LCN3 B8YLY6 Q9C739 Q9LH44 A0A2N9GX95 A0A2N9GHK9 A0A0N1ICY1 A0A151QNH5 A0A151RJN4 A0A151TMN9 A0A224XJ98 B8YLY4 A0A151SDY6 B8YLY3 A0A059QBK0 Q9SXB2 A0A1Y1M2G6 A0A0A9XEU3 A0A151R6U8 Q9C536 A0A194RJV9 A0A0J7K6W1 B8YLY5 Q9M2D1 A0A0V0RWJ3 A0A2Z6NRJ0 A0A2N9GHB5 A0A2N9F1W0 A0A151RAL1 A0A194RU38 A0A034VP82 A0A087SVE4 A0A151R2G6 A0A2A4J959 A0A131XNF9 A0A2R6RYD4 A0A1Y1MYL8 A0A151QZS0 A0A0P6K133 A0A0P6J144 A0A151TKT1 A0A061F3M7 A0A1Y1MYL6 A0A151T469 A0A0V0TUP0 A0A2K3N1R3 A0A2N9ESI4 A0A2N9J3Y8 E5SJ15 A0A151RBY1 E5SZD6 Q6L3N8 A0A1W7R685 A0A2N9HHD8 A0A151S6I6 E5SB91 A0A2K3MZ63 A0A2N9I1L5 A0A2N9FNY4 A0A151TIH6 A0A0V1APS7 A0A151SHZ9
PDB
3S3N
E-value=2.59438e-07,
Score=136
Ontologies
KEGG
GO
PANTHER
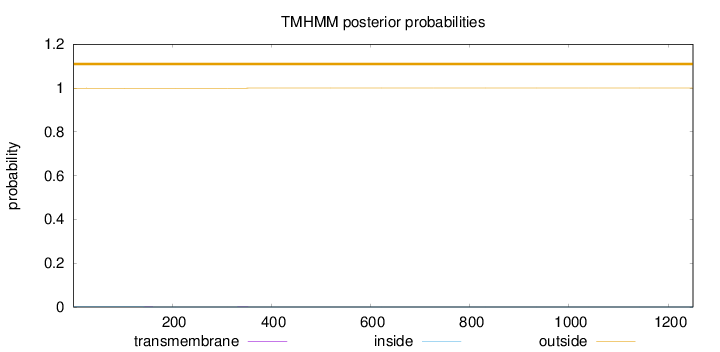
Topology
Length:
1250
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0131
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.00061
outside
1 - 1250
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
-0.791117
CLR
3.292376
CSRT
0
Interpretation
Uncertain
