Gene
KWMTBOMO00011
Pre Gene Modal
BGIBMGA002076
Annotation
PREDICTED:_SET_and_MYND_domain-containing_protein_4_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.999
Sequence
CDS
ATGTCTCGAATTTACGAAGAAGTGGATCCCATATACGCTGCGACCTGTAGCGACATCACTCTTTGCTCAAATTCCAAAGGTTTCTTCAAGGGTCTCGCCGATGACCTGGTTTCATTAGCTGGGGAAGAATGGCTAAATAAATTTGAGCTAGTGGAAGATGGCAAGAAAGTTACATTTTTTATGGAGAATAAGGAGGTAATGGAAGCACTTACTGAAGTATTATCCAGGATACAGCCCCTTCACCGCGGGAAAGACGCTAGAATATCGAGTCAGAGGCGCGGCGACGCACAATCGGCTCTGAAGAATGAAGATCTAATGAAGGCGTTGGCGTTGGCAAGTCAAGCCGTACTCAGGGCTCCCGTTACGGGAGAAAATGAAGCGATTGACGGCGGGGTGTCGCTGGCTTTGGCGCTGTGGCTTCGGTCTGAGATCCTGCTGAAATTAAATAGACCTCAAGCCGCCCTAGAAGATTTGAAGTTAGCTCTAAAGGAACGCCTACCTGCCAGAATGCGAGCCCACTACTACTGGAGAATGGGACACTGCTACCGAGGTACCGGAGAGGCTACAAGAGCGAAGGTGTCGTATGAACTGGCCGGCCGACTGCTCGCTAAGGACGAGGTCGCCAAAACACAGCTAACTAAAGACATTGAAACTCTAGATTATACAGTACAATCGAAACGGCCGCCCGATAAGAATGCCACCACACTAACCGGAGGAGCGAAATTAAACTTGCCATCTTTATCGAAACTACTGAAAATAGTAGAAGAAGAGAACAAGGGTCGCTTCGCCGTGGCCAGTGCTCCGGTTCGAACGGGAGACGTCCTGCTGGTTGACAGTCCGTACGCAGCCTGCCTGCTCTCGGATTACTACGGAACGCACTGCCTTCATTGTTTCAGAAGGTTGGCGGACTGCGAAGAGTCGGCCCCGGTGTGGTGTCCCAAATGCTCAGGCGTTGTGTTCTGTAGTATCGAATGTCGCGATACTGCGGTTTCTTCGTATCACTCCTTTGAGTGCCAGTTCCTAGATCTATTTGTGGGTTCCGGAATGTCCATCCTGAGTCACATTGCTCTCAGAATGGTGACACAATCAGATCTCGAAACGTGTCTGACAATCCACTCAAAGTACATCTCCAATGACATTAAAACCGTTGAAGGGAGCGTGCTTAACGACATCGAAGGCGTTGCCAAAAAATCTAAAATGAAAAGTAGAAAAGAGAGACTGAATAGAAATAAGAAAAGCCATAAGATGTCTGTAGAGAAGAATGATAGAGTTGACGTAATGGAAGATAAACTGGAAGACAAAAATAATTTTGAAGAGAAGTTAGAATTGAAAGCGGCGCAAGTTTACTCTCTCTGTACACACTCTGACCGGAGAAGGGGTGATGATTACTTGAAGAGGATAGTGATGGGCTATTTCCTGACAGAATGCCTGAAGCACGCTGGATTTTTCAAAAACTGCAATAAAAATAATTTGACTAAAGCGCAACAGTCCATATGTGAACTGATAGTTCGCAACCTGCAGTTGTTACAATTCAACGCACACGAAATTTACGAGACCGTGCGAGGAGAGCACCAGTTCAGCGGGTCCAAGCCGCTCTATTTCGCTGTGGGGATCTACCCTGTCGGAGCACTCTTCAACCACGAGTGCTACCCCGCGGTCACCAGATATTTTGAGGGTCGTAAGATCGTCCTACGCGCAACGAGACCCCTGACTCCCGGAGAAGTGGTCTCCGAAAATTACGGACCGCACTTCATGATGCGGACCCTGAGGGAAAGGCAGCGAGCCCTCGCCTGCCGGTACTGGTTCCACTGTGAATGTACCGCTTGTAAGGAAGACTGGCCAACTATGAAGCAGATGAATAACGATTCTATTTCTTACATAAGATGTTCTAACCTCGCCTGCCGAGGTAAGCTCCGAGGGAGTGTCCAGCGGATGGGTGATCGATGCAGCCTATGCTCTACCCCTATAGACAAGGATCTAGTAACAGTTAAAATTGACACGATCAATAAATGCACGGCTCAGTATCAGGAAGGCGCCAAGTTAATGGACAAAGAGATGCCAGAAGAAGCCACCGCGACACTGTGCAGTGCTATAGACTCGTTTCACGAGGCAGGCCGGCCTCCTCACCTTGAAACGCACTTAGCCCAAGAAGCCTTAAGATCATGTTTTGCAGTACGTGGAAACATTCACATCCTGAAGAAGGATGGCGAAAAAGACAACGAAAACAAGTAA
Protein
MSRIYEEVDPIYAATCSDITLCSNSKGFFKGLADDLVSLAGEEWLNKFELVEDGKKVTFFMENKEVMEALTEVLSRIQPLHRGKDARISSQRRGDAQSALKNEDLMKALALASQAVLRAPVTGENEAIDGGVSLALALWLRSEILLKLNRPQAALEDLKLALKERLPARMRAHYYWRMGHCYRGTGEATRAKVSYELAGRLLAKDEVAKTQLTKDIETLDYTVQSKRPPDKNATTLTGGAKLNLPSLSKLLKIVEEENKGRFAVASAPVRTGDVLLVDSPYAACLLSDYYGTHCLHCFRRLADCEESAPVWCPKCSGVVFCSIECRDTAVSSYHSFECQFLDLFVGSGMSILSHIALRMVTQSDLETCLTIHSKYISNDIKTVEGSVLNDIEGVAKKSKMKSRKERLNRNKKSHKMSVEKNDRVDVMEDKLEDKNNFEEKLELKAAQVYSLCTHSDRRRGDDYLKRIVMGYFLTECLKHAGFFKNCNKNNLTKAQQSICELIVRNLQLLQFNAHEIYETVRGEHQFSGSKPLYFAVGIYPVGALFNHECYPAVTRYFEGRKIVLRATRPLTPGEVVSENYGPHFMMRTLRERQRALACRYWFHCECTACKEDWPTMKQMNNDSISYIRCSNLACRGKLRGSVQRMGDRCSLCSTPIDKDLVTVKIDTINKCTAQYQEGAKLMDKEMPEEATATLCSAIDSFHEAGRPPHLETHLAQEALRSCFAVRGNIHILKKDGEKDNENK
Summary
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Uniprot
H9IXU3
A0A2A4JBJ6
A0A212EIB3
A0A194QXS2
A0A194PLY3
A0A2P8YCF3
+ More
A0A088ARS2 A0A3L8DG49 A0A158NS58 A0A336KP35 A0A0C9Q6Y6 A0A336MH96 E0VRD3 A0A154PBQ3 A0A1B6DV01 A0A1B6DJ26 A0A067R1I2 A0A0P6JC10 A0A0N7ZM19 A0A026WYK2 A0A1J1J3W6 A0A0L7RDH1 A0A195BZ02 A0A0N8BLB7 A0A0P4YU54 A0A0P5K0D5 A0A0N8BSQ5 A0A0P6H8T4 A0A0P4W687 A0A0P4W8Y7 A0A023F0G4 Q0IG30 A0A1S4F4X7 A0A0K2TLD8 J9JK42 E9I266 A0A1B6CWC9 A0A0P5ZB00 A0A0N8B7W5 B0X7W3 A0A0P5AZF7 A0A0P6BIH0 A0A0P5XZ86 A0A0P6GEJ2 A0A0P5NTU3 A0A0P5HLE6 A0A0P5S7Y6 A0A0P5NL24 A0A0P5S3C9
A0A088ARS2 A0A3L8DG49 A0A158NS58 A0A336KP35 A0A0C9Q6Y6 A0A336MH96 E0VRD3 A0A154PBQ3 A0A1B6DV01 A0A1B6DJ26 A0A067R1I2 A0A0P6JC10 A0A0N7ZM19 A0A026WYK2 A0A1J1J3W6 A0A0L7RDH1 A0A195BZ02 A0A0N8BLB7 A0A0P4YU54 A0A0P5K0D5 A0A0N8BSQ5 A0A0P6H8T4 A0A0P4W687 A0A0P4W8Y7 A0A023F0G4 Q0IG30 A0A1S4F4X7 A0A0K2TLD8 J9JK42 E9I266 A0A1B6CWC9 A0A0P5ZB00 A0A0N8B7W5 B0X7W3 A0A0P5AZF7 A0A0P6BIH0 A0A0P5XZ86 A0A0P6GEJ2 A0A0P5NTU3 A0A0P5HLE6 A0A0P5S7Y6 A0A0P5NL24 A0A0P5S3C9
Pubmed
EMBL
BABH01015606
BABH01015607
NWSH01002090
PCG69219.1
AGBW02014676
OWR41217.1
+ More
KQ460953 KPJ10338.1 KQ459599 KPI94337.1 PYGN01000705 PSN41945.1 QOIP01000008 RLU19281.1 ADTU01024480 ADTU01024481 ADTU01024482 ADTU01024483 ADTU01024484 ADTU01024485 UFQS01000586 UFQT01000586 SSX05155.1 SSX25516.1 GBYB01009883 JAG79650.1 UFQT01000990 SSX28319.1 DS235465 EEB15939.1 KQ434856 KZC08688.1 GEDC01007782 JAS29516.1 GEDC01011594 JAS25704.1 KK853097 KDR11443.1 GDIQ01009517 JAN85220.1 GDIP01232185 JAI91216.1 KK107064 EZA60928.1 CVRI01000067 CRL06484.1 KQ414614 KOC68811.1 KQ978457 KYM93842.1 GDIQ01166523 JAK85202.1 GDIP01233322 JAI90079.1 GDIQ01190994 JAK60731.1 GDIQ01148619 JAL03107.1 GDIQ01035071 JAN59666.1 GDRN01080085 JAI62291.1 GDRN01080086 GDRN01080084 GDRN01080083 JAI62293.1 GBBI01003755 JAC14957.1 CH477274 EAT45183.1 HACA01009497 CDW26858.1 ABLF02026503 GL733993 EFX61914.1 GEDC01019561 JAS17737.1 GDIP01047248 JAM56467.1 GDIQ01204139 JAK47586.1 DS232465 EDS42182.1 GDIP01192235 JAJ31167.1 GDIP01014570 JAM89145.1 GDIP01065300 JAM38415.1 GDIQ01044758 JAN49979.1 GDIQ01148031 JAL03695.1 GDIQ01231737 JAK19988.1 GDIQ01091286 JAL60440.1 GDIQ01147980 JAL03746.1 GDIQ01093574 JAL58152.1
KQ460953 KPJ10338.1 KQ459599 KPI94337.1 PYGN01000705 PSN41945.1 QOIP01000008 RLU19281.1 ADTU01024480 ADTU01024481 ADTU01024482 ADTU01024483 ADTU01024484 ADTU01024485 UFQS01000586 UFQT01000586 SSX05155.1 SSX25516.1 GBYB01009883 JAG79650.1 UFQT01000990 SSX28319.1 DS235465 EEB15939.1 KQ434856 KZC08688.1 GEDC01007782 JAS29516.1 GEDC01011594 JAS25704.1 KK853097 KDR11443.1 GDIQ01009517 JAN85220.1 GDIP01232185 JAI91216.1 KK107064 EZA60928.1 CVRI01000067 CRL06484.1 KQ414614 KOC68811.1 KQ978457 KYM93842.1 GDIQ01166523 JAK85202.1 GDIP01233322 JAI90079.1 GDIQ01190994 JAK60731.1 GDIQ01148619 JAL03107.1 GDIQ01035071 JAN59666.1 GDRN01080085 JAI62291.1 GDRN01080086 GDRN01080084 GDRN01080083 JAI62293.1 GBBI01003755 JAC14957.1 CH477274 EAT45183.1 HACA01009497 CDW26858.1 ABLF02026503 GL733993 EFX61914.1 GEDC01019561 JAS17737.1 GDIP01047248 JAM56467.1 GDIQ01204139 JAK47586.1 DS232465 EDS42182.1 GDIP01192235 JAJ31167.1 GDIP01014570 JAM89145.1 GDIP01065300 JAM38415.1 GDIQ01044758 JAN49979.1 GDIQ01148031 JAL03695.1 GDIQ01231737 JAK19988.1 GDIQ01091286 JAL60440.1 GDIQ01147980 JAL03746.1 GDIQ01093574 JAL58152.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IXU3
A0A2A4JBJ6
A0A212EIB3
A0A194QXS2
A0A194PLY3
A0A2P8YCF3
+ More
A0A088ARS2 A0A3L8DG49 A0A158NS58 A0A336KP35 A0A0C9Q6Y6 A0A336MH96 E0VRD3 A0A154PBQ3 A0A1B6DV01 A0A1B6DJ26 A0A067R1I2 A0A0P6JC10 A0A0N7ZM19 A0A026WYK2 A0A1J1J3W6 A0A0L7RDH1 A0A195BZ02 A0A0N8BLB7 A0A0P4YU54 A0A0P5K0D5 A0A0N8BSQ5 A0A0P6H8T4 A0A0P4W687 A0A0P4W8Y7 A0A023F0G4 Q0IG30 A0A1S4F4X7 A0A0K2TLD8 J9JK42 E9I266 A0A1B6CWC9 A0A0P5ZB00 A0A0N8B7W5 B0X7W3 A0A0P5AZF7 A0A0P6BIH0 A0A0P5XZ86 A0A0P6GEJ2 A0A0P5NTU3 A0A0P5HLE6 A0A0P5S7Y6 A0A0P5NL24 A0A0P5S3C9
A0A088ARS2 A0A3L8DG49 A0A158NS58 A0A336KP35 A0A0C9Q6Y6 A0A336MH96 E0VRD3 A0A154PBQ3 A0A1B6DV01 A0A1B6DJ26 A0A067R1I2 A0A0P6JC10 A0A0N7ZM19 A0A026WYK2 A0A1J1J3W6 A0A0L7RDH1 A0A195BZ02 A0A0N8BLB7 A0A0P4YU54 A0A0P5K0D5 A0A0N8BSQ5 A0A0P6H8T4 A0A0P4W687 A0A0P4W8Y7 A0A023F0G4 Q0IG30 A0A1S4F4X7 A0A0K2TLD8 J9JK42 E9I266 A0A1B6CWC9 A0A0P5ZB00 A0A0N8B7W5 B0X7W3 A0A0P5AZF7 A0A0P6BIH0 A0A0P5XZ86 A0A0P6GEJ2 A0A0P5NTU3 A0A0P5HLE6 A0A0P5S7Y6 A0A0P5NL24 A0A0P5S3C9
PDB
3OXG
E-value=1.49629e-06,
Score=127
Ontologies
GO
Topology
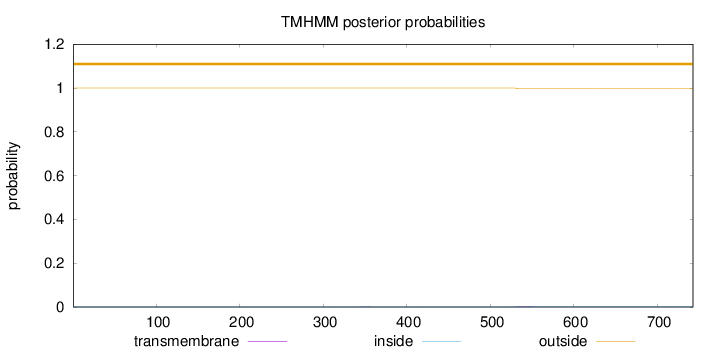
Length:
743
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04909
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00027
outside
1 - 743
Population Genetic Test Statistics
Pi
23.560094
Theta
20.88344
Tajima's D
-1.903412
CLR
0.015593
CSRT
0.0184990750462477
Interpretation
Uncertain
