Gene
KWMTBOMO00005
Pre Gene Modal
BGIBMGA002069
Annotation
PREDICTED:_tyrosine-protein_kinase_Abl_isoform_X1_[Bombyx_mori]
Full name
Tyrosine-protein kinase
+ More
Non-specific protein-tyrosine kinase
Non-specific protein-tyrosine kinase
Location in the cell
Mitochondrial Reliability : 1.919
Sequence
CDS
ATGGGCGCGCAGCAGGCCAAAGAACGCGGCACCGCCAGCGGCGCGTCCATGCGTTCGGCGCGCAGCAAGCCACGGGTGCCGAAGGACCCCCGCATGCTCGGCTCCAACATATTCACTGAACATAGCGGGTGTGTCTCAGCGTGGAGATGGACCTCGAAGGAGAATCTTTTGGCCCATCACGAAGAGGACGATCCCCAGCTGTTCGTAGCCCTCTACGATTTCCAGGCGGGAGGGGAGAACCAGCTCACGCTCAAGAAAGGCGAGCAGGTGCGCATCATGAGCTACAACAAGAGCGGCGAGTGGTGCGAAGCCCACACGCTGACTGGGGCCGTGGGCTGGGTGCCCAGCAACTACGTCACTCCAGTCAACAGTCTCGAGAAGCATTCATGGTACCATGGGCCGATTTCCCGCAACGCGGCGGAGTACCTCCTCTCGTCTGGGATCAATGGCAGCTTCCTGGTCCGCGAATCGGAATCGAGTCCTGGGCAACGCAGCATATCCCTCCGCTACGAAGGGAGAGTTTACCACTACCGCATCAACGAAGACGCCGATGGGAAGGTTTATGTGACCTCCGAGTCGAAGTTCGGAACGTTAGCGGAGCTGGTCCACCATCACTCGGTGGCGGGGGACGGTCTAATTACGCAGCTGCTGTATCCAGCACCGAAGCGCTCGAAGCCTACAGTGTTCCCCCTGGCTCCCCCCGACCACTGGGAGATCGATCGCACGGACATAGTGATGAAGCACAAGCTGGGCGGCGGCCAGTATGGAGACGTTTACGAAGCCGCTTGGAAGCGCGGCAACATCACGGTGGCGGTCAAAACACTCAAGGACGATACGATGGCGCTCAAGGATTTCCTGGAGGAAGCCTCCATAATGAAAGAGATGCGACACCCGAACCTGGTGCAGCTGCTCGGCGTGTGCACACGCGAGCCCCCGTTCTATATTATTACGGAGTTTATGTCCCGCGGAAACCTGCTGGAGTACCTGCGGGCGGGGGCGAGGGAGTGCGTGCCGGGCGCGGTCGTGCTCATGTACATGGCGACGCAGATAGCGTCCGGCATGAGCTACCTCGAGAGCCGCTCCTTCATCCACCGCGACCTCGCCGCCAGGAACTGCCTCGTGGGGGAGAACCATCTGGTCAAGGTGGCCGACTTCGGACTGGCTCGCCTGATGCGCGACGACACATACACGGCCCACGCCGGCGCCAAGTTCCCGATCAAGTGGACGGCGCCGGAGGGTCTGGCCTACAACACTTTCAGCACCAAATCGGATGTCTGGGCTTTCGGGATCCTGCTGTGGGAGATCGCCACGTACGGGATGTCTCCTTACCCCGGAGTGGATCTCGCGGACGTTTACCACATGCTTGAGAAGGGGTACCGCATGGAGTGTCCCCCGGGCTGCCCGGCGCCCGTCTACGAGCTCATGCGCGGCTGCTGGCAGTGGAGCCCCTCCGAGCGGCCCTCCTTCCGCGAGATACACCACGCGCTCGAGCACATGTTCCAGGACAACTCGATCACAGACGAAGTAGAGAAACAACTCCAAGAAGGCAGCGGCGCGCAGGCCGCCGGCACCCCGCTGCTGTCGCTGAAGAAGACGAGCGCCGACCCTCGAGCGGTGCAGATGCGGCGCCCGACCAACCGCCGCGGCAAGCAAGCCCCCACCCCTCCCAAACGCACTAGCCTGCTGTCCTCGTGCAGCTCCTTCCGCGAGTCGCAGTACGCCGCCGACGAGCACGCGCCCGACGACGCGCCCGCCGACGCGCTCGCCGACCTCAACGGTCAGTGTCTCACGCAGCGTCACGCGCCGTCACGCACTCCAACAACCGATATAGTGCAATAG
Protein
MGAQQAKERGTASGASMRSARSKPRVPKDPRMLGSNIFTEHSGCVSAWRWTSKENLLAHHEEDDPQLFVALYDFQAGGENQLTLKKGEQVRIMSYNKSGEWCEAHTLTGAVGWVPSNYVTPVNSLEKHSWYHGPISRNAAEYLLSSGINGSFLVRESESSPGQRSISLRYEGRVYHYRINEDADGKVYVTSESKFGTLAELVHHHSVAGDGLITQLLYPAPKRSKPTVFPLAPPDHWEIDRTDIVMKHKLGGGQYGDVYEAAWKRGNITVAVKTLKDDTMALKDFLEEASIMKEMRHPNLVQLLGVCTREPPFYIITEFMSRGNLLEYLRAGARECVPGAVVLMYMATQIASGMSYLESRSFIHRDLAARNCLVGENHLVKVADFGLARLMRDDTYTAHAGAKFPIKWTAPEGLAYNTFSTKSDVWAFGILLWEIATYGMSPYPGVDLADVYHMLEKGYRMECPPGCPAPVYELMRGCWQWSPSERPSFREIHHALEHMFQDNSITDEVEKQLQEGSGAQAAGTPLLSLKKTSADPRAVQMRRPTNRRGKQAPTPPKRTSLLSSCSSFRESQYAADEHAPDDAPADALADLNGQCLTQRHAPSRTPTTDIVQ
Summary
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Uniprot
A0A2W1BXZ4
A0A212EXN4
A0A194QLS4
A0A194PFC7
A0A1B6CDF7
A0A1B6DYH9
+ More
A0A1W4WJG9 A0A1W4WUB1 A0A1W4WVJ8 A0A1B6HCF1 A0A139WK02 A0A1Y1KNK4 A0A1Y1KMS3 A0A1Y1KK61 A0A1Y1KKE9 A0A1B6JBG0 A0A0L7R834 A0A139WJS9 A0A1Y1KQR3 A0A1Y1KK43 A0A1Y1KQS3 A0A1Y1KK48 A0A3L8DM09 A0A026W235 A0A0C9PHZ8 A0A0P4VT99 A0A0M9AB24 A0A088AT87 A0A069DY91 A0A224X5G0 A0A2A3EUP4 F4WV60 K7J766 A0A158NUM9 A0A151WUC9 A0A195DS06 A0A195EXC2 A0A232ES62 A0A195C704 J9K2P5 A0A1W4WJI7 A0A1Y1KK51 A0A1Y1KK32 A0A1Y1KNJ1 A0A1W4WVB9 A0A195BS54 A0A1Y1KQ07 A0A067RM58 A0A1Y1KK29 A0A0K8SX23 A0A0K8TEK1 A0A1Y1KSD4 A0A0J7L3V4 A0A1Y1KQ17 A0A1Y1KKC9 A0A1Y1KQ27 A0A0K8SVW5 A0A1Y1KMH7 A0A1B6D953 A0A1B6DM49 A0A1B6EB12 A0A1B6C175 A0A1Y1KK27 E0VZT3 T1HWB4 A0A1Y1KK49 A0A336KXW8 A0A1Q3F495 A0A146KZU4 A0A0K8TF41 A0A0K8T914 A0A0K8SVV8 A0A0K8TET8 A0A1W7R720 Q16XS6 A0A1S4FKD3 A0A2S2P7F6 A0A0K8SVW2 A0A2M3Z2A4 W4VRS5 B0WAD3 A0A336MRU8 A0A1I8N9R0 A0A1I8N9R1 A0A1I8N9Q8 A0A1I8NNX1 A0A1I8N9S0 A0A1B0BMW2 A0A146LW71 A0A1I8NNX9 A0A1I8NNX8 A0A1I8NNX3 A0A1I8NNZ3 A0A336LL25 A0A0A1X0T2 A0A2M4BFH1 W5J694 A0A2M3Z6K3 A0A2M4CG35 A0A2M4AMJ2
A0A1W4WJG9 A0A1W4WUB1 A0A1W4WVJ8 A0A1B6HCF1 A0A139WK02 A0A1Y1KNK4 A0A1Y1KMS3 A0A1Y1KK61 A0A1Y1KKE9 A0A1B6JBG0 A0A0L7R834 A0A139WJS9 A0A1Y1KQR3 A0A1Y1KK43 A0A1Y1KQS3 A0A1Y1KK48 A0A3L8DM09 A0A026W235 A0A0C9PHZ8 A0A0P4VT99 A0A0M9AB24 A0A088AT87 A0A069DY91 A0A224X5G0 A0A2A3EUP4 F4WV60 K7J766 A0A158NUM9 A0A151WUC9 A0A195DS06 A0A195EXC2 A0A232ES62 A0A195C704 J9K2P5 A0A1W4WJI7 A0A1Y1KK51 A0A1Y1KK32 A0A1Y1KNJ1 A0A1W4WVB9 A0A195BS54 A0A1Y1KQ07 A0A067RM58 A0A1Y1KK29 A0A0K8SX23 A0A0K8TEK1 A0A1Y1KSD4 A0A0J7L3V4 A0A1Y1KQ17 A0A1Y1KKC9 A0A1Y1KQ27 A0A0K8SVW5 A0A1Y1KMH7 A0A1B6D953 A0A1B6DM49 A0A1B6EB12 A0A1B6C175 A0A1Y1KK27 E0VZT3 T1HWB4 A0A1Y1KK49 A0A336KXW8 A0A1Q3F495 A0A146KZU4 A0A0K8TF41 A0A0K8T914 A0A0K8SVV8 A0A0K8TET8 A0A1W7R720 Q16XS6 A0A1S4FKD3 A0A2S2P7F6 A0A0K8SVW2 A0A2M3Z2A4 W4VRS5 B0WAD3 A0A336MRU8 A0A1I8N9R0 A0A1I8N9R1 A0A1I8N9Q8 A0A1I8NNX1 A0A1I8N9S0 A0A1B0BMW2 A0A146LW71 A0A1I8NNX9 A0A1I8NNX8 A0A1I8NNX3 A0A1I8NNZ3 A0A336LL25 A0A0A1X0T2 A0A2M4BFH1 W5J694 A0A2M3Z6K3 A0A2M4CG35 A0A2M4AMJ2
EC Number
2.7.10.2
Pubmed
EMBL
KZ149922
PZC77690.1
AGBW02011730
OWR46217.1
KQ461196
KPJ06314.1
+ More
KQ459605 KPI91987.1 GEDC01025811 JAS11487.1 GEDC01006577 JAS30721.1 GECU01035414 JAS72292.1 KQ971331 KYB28309.1 GEZM01081231 GEZM01081216 JAV61780.1 GEZM01081222 GEZM01081217 JAV61771.1 GEZM01081225 GEZM01081206 JAV61793.1 GEZM01081215 GEZM01081205 JAV61794.1 GECU01011192 JAS96514.1 KQ414637 KOC67004.1 KYB28308.1 GEZM01081228 GEZM01081218 JAV61776.1 GEZM01081226 GEZM01081221 JAV61773.1 GEZM01081211 GEZM01081207 JAV61786.1 GEZM01081232 GEZM01081213 JAV61783.1 QOIP01000006 RLU21464.1 KK107485 EZA50127.1 GBYB01000512 GBYB01000513 GBYB01000515 JAG70279.1 JAG70280.1 JAG70282.1 GDKW01000146 JAI56449.1 KQ435694 KOX80895.1 GBGD01000157 JAC88732.1 GFTR01008691 JAW07735.1 KZ288185 PBC34936.1 GL888384 EGI61948.1 AAZX01007534 ADTU01026625 KQ982745 KYQ51367.1 KQ980530 KYN15631.1 KQ981953 KYN32529.1 NNAY01002531 OXU21126.1 KQ978164 KYM96644.1 ABLF02012147 GEZM01081208 JAV61789.1 GEZM01081224 JAV61769.1 GEZM01081223 JAV61770.1 KQ976417 KYM89862.1 GEZM01081227 JAV61765.1 KK852591 KDR20715.1 GEZM01081229 JAV61763.1 GBRD01008376 JAG57445.1 GBRD01001829 JAG63992.1 GEZM01081210 JAV61787.1 LBMM01000867 KMQ97331.1 GEZM01081219 JAV61775.1 GEZM01081220 JAV61774.1 GEZM01081212 JAV61785.1 GBRD01008374 JAG57447.1 GEZM01081209 JAV61788.1 GEDC01015116 JAS22182.1 GEDC01010576 JAS26722.1 GEDC01002162 JAS35136.1 GEDC01030279 JAS07019.1 GEZM01081230 JAV61762.1 DS235854 EEB18889.1 ACPB03013644 ACPB03013645 GEZM01081214 JAV61782.1 UFQS01001061 UFQT01001061 SSX08755.1 SSX28667.1 GFDL01012727 JAV22318.1 GDHC01018409 JAQ00220.1 GBRD01001830 JAG63991.1 GBRD01003789 JAG62032.1 GBRD01008383 JAG57438.1 GBRD01001827 JAG63994.1 GEHC01000725 JAV46920.1 CH477533 EAT39429.1 GGMR01012772 MBY25391.1 GBRD01008378 JAG57443.1 GGFM01001817 MBW22568.1 GANO01000206 JAB59665.1 DS231870 EDS41023.1 SSX08756.1 SSX28668.1 JXJN01017066 GDHC01006736 JAQ11893.1 UFQS01000040 UFQT01000040 SSW98280.1 SSX18666.1 GBXI01009937 JAD04355.1 GGFJ01002659 MBW51800.1 ADMH02002077 ETN59521.1 GGFM01003405 MBW24156.1 GGFL01000128 MBW64306.1 GGFK01008694 MBW42015.1
KQ459605 KPI91987.1 GEDC01025811 JAS11487.1 GEDC01006577 JAS30721.1 GECU01035414 JAS72292.1 KQ971331 KYB28309.1 GEZM01081231 GEZM01081216 JAV61780.1 GEZM01081222 GEZM01081217 JAV61771.1 GEZM01081225 GEZM01081206 JAV61793.1 GEZM01081215 GEZM01081205 JAV61794.1 GECU01011192 JAS96514.1 KQ414637 KOC67004.1 KYB28308.1 GEZM01081228 GEZM01081218 JAV61776.1 GEZM01081226 GEZM01081221 JAV61773.1 GEZM01081211 GEZM01081207 JAV61786.1 GEZM01081232 GEZM01081213 JAV61783.1 QOIP01000006 RLU21464.1 KK107485 EZA50127.1 GBYB01000512 GBYB01000513 GBYB01000515 JAG70279.1 JAG70280.1 JAG70282.1 GDKW01000146 JAI56449.1 KQ435694 KOX80895.1 GBGD01000157 JAC88732.1 GFTR01008691 JAW07735.1 KZ288185 PBC34936.1 GL888384 EGI61948.1 AAZX01007534 ADTU01026625 KQ982745 KYQ51367.1 KQ980530 KYN15631.1 KQ981953 KYN32529.1 NNAY01002531 OXU21126.1 KQ978164 KYM96644.1 ABLF02012147 GEZM01081208 JAV61789.1 GEZM01081224 JAV61769.1 GEZM01081223 JAV61770.1 KQ976417 KYM89862.1 GEZM01081227 JAV61765.1 KK852591 KDR20715.1 GEZM01081229 JAV61763.1 GBRD01008376 JAG57445.1 GBRD01001829 JAG63992.1 GEZM01081210 JAV61787.1 LBMM01000867 KMQ97331.1 GEZM01081219 JAV61775.1 GEZM01081220 JAV61774.1 GEZM01081212 JAV61785.1 GBRD01008374 JAG57447.1 GEZM01081209 JAV61788.1 GEDC01015116 JAS22182.1 GEDC01010576 JAS26722.1 GEDC01002162 JAS35136.1 GEDC01030279 JAS07019.1 GEZM01081230 JAV61762.1 DS235854 EEB18889.1 ACPB03013644 ACPB03013645 GEZM01081214 JAV61782.1 UFQS01001061 UFQT01001061 SSX08755.1 SSX28667.1 GFDL01012727 JAV22318.1 GDHC01018409 JAQ00220.1 GBRD01001830 JAG63991.1 GBRD01003789 JAG62032.1 GBRD01008383 JAG57438.1 GBRD01001827 JAG63994.1 GEHC01000725 JAV46920.1 CH477533 EAT39429.1 GGMR01012772 MBY25391.1 GBRD01008378 JAG57443.1 GGFM01001817 MBW22568.1 GANO01000206 JAB59665.1 DS231870 EDS41023.1 SSX08756.1 SSX28668.1 JXJN01017066 GDHC01006736 JAQ11893.1 UFQS01000040 UFQT01000040 SSW98280.1 SSX18666.1 GBXI01009937 JAD04355.1 GGFJ01002659 MBW51800.1 ADMH02002077 ETN59521.1 GGFM01003405 MBW24156.1 GGFL01000128 MBW64306.1 GGFK01008694 MBW42015.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000192223
UP000007266
UP000053825
+ More
UP000279307 UP000053097 UP000053105 UP000005203 UP000242457 UP000007755 UP000002358 UP000005205 UP000075809 UP000078492 UP000078541 UP000215335 UP000078542 UP000007819 UP000078540 UP000027135 UP000036403 UP000009046 UP000015103 UP000008820 UP000002320 UP000095301 UP000095300 UP000092460 UP000000673
UP000279307 UP000053097 UP000053105 UP000005203 UP000242457 UP000007755 UP000002358 UP000005205 UP000075809 UP000078492 UP000078541 UP000215335 UP000078542 UP000007819 UP000078540 UP000027135 UP000036403 UP000009046 UP000015103 UP000008820 UP000002320 UP000095301 UP000095300 UP000092460 UP000000673
Interpro
IPR008266
Tyr_kinase_AS
+ More
IPR035837 ABL_SH2
IPR036860 SH2_dom_sf
IPR033221 ABL1
IPR036028 SH3-like_dom_sf
IPR000719 Prot_kinase_dom
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR001452 SH3_domain
IPR000980 SH2
IPR020635 Tyr_kinase_cat_dom
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR015015 F-actin_binding
IPR035837 ABL_SH2
IPR036860 SH2_dom_sf
IPR033221 ABL1
IPR036028 SH3-like_dom_sf
IPR000719 Prot_kinase_dom
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR001452 SH3_domain
IPR000980 SH2
IPR020635 Tyr_kinase_cat_dom
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR015015 F-actin_binding
Gene 3D
CDD
ProteinModelPortal
A0A2W1BXZ4
A0A212EXN4
A0A194QLS4
A0A194PFC7
A0A1B6CDF7
A0A1B6DYH9
+ More
A0A1W4WJG9 A0A1W4WUB1 A0A1W4WVJ8 A0A1B6HCF1 A0A139WK02 A0A1Y1KNK4 A0A1Y1KMS3 A0A1Y1KK61 A0A1Y1KKE9 A0A1B6JBG0 A0A0L7R834 A0A139WJS9 A0A1Y1KQR3 A0A1Y1KK43 A0A1Y1KQS3 A0A1Y1KK48 A0A3L8DM09 A0A026W235 A0A0C9PHZ8 A0A0P4VT99 A0A0M9AB24 A0A088AT87 A0A069DY91 A0A224X5G0 A0A2A3EUP4 F4WV60 K7J766 A0A158NUM9 A0A151WUC9 A0A195DS06 A0A195EXC2 A0A232ES62 A0A195C704 J9K2P5 A0A1W4WJI7 A0A1Y1KK51 A0A1Y1KK32 A0A1Y1KNJ1 A0A1W4WVB9 A0A195BS54 A0A1Y1KQ07 A0A067RM58 A0A1Y1KK29 A0A0K8SX23 A0A0K8TEK1 A0A1Y1KSD4 A0A0J7L3V4 A0A1Y1KQ17 A0A1Y1KKC9 A0A1Y1KQ27 A0A0K8SVW5 A0A1Y1KMH7 A0A1B6D953 A0A1B6DM49 A0A1B6EB12 A0A1B6C175 A0A1Y1KK27 E0VZT3 T1HWB4 A0A1Y1KK49 A0A336KXW8 A0A1Q3F495 A0A146KZU4 A0A0K8TF41 A0A0K8T914 A0A0K8SVV8 A0A0K8TET8 A0A1W7R720 Q16XS6 A0A1S4FKD3 A0A2S2P7F6 A0A0K8SVW2 A0A2M3Z2A4 W4VRS5 B0WAD3 A0A336MRU8 A0A1I8N9R0 A0A1I8N9R1 A0A1I8N9Q8 A0A1I8NNX1 A0A1I8N9S0 A0A1B0BMW2 A0A146LW71 A0A1I8NNX9 A0A1I8NNX8 A0A1I8NNX3 A0A1I8NNZ3 A0A336LL25 A0A0A1X0T2 A0A2M4BFH1 W5J694 A0A2M3Z6K3 A0A2M4CG35 A0A2M4AMJ2
A0A1W4WJG9 A0A1W4WUB1 A0A1W4WVJ8 A0A1B6HCF1 A0A139WK02 A0A1Y1KNK4 A0A1Y1KMS3 A0A1Y1KK61 A0A1Y1KKE9 A0A1B6JBG0 A0A0L7R834 A0A139WJS9 A0A1Y1KQR3 A0A1Y1KK43 A0A1Y1KQS3 A0A1Y1KK48 A0A3L8DM09 A0A026W235 A0A0C9PHZ8 A0A0P4VT99 A0A0M9AB24 A0A088AT87 A0A069DY91 A0A224X5G0 A0A2A3EUP4 F4WV60 K7J766 A0A158NUM9 A0A151WUC9 A0A195DS06 A0A195EXC2 A0A232ES62 A0A195C704 J9K2P5 A0A1W4WJI7 A0A1Y1KK51 A0A1Y1KK32 A0A1Y1KNJ1 A0A1W4WVB9 A0A195BS54 A0A1Y1KQ07 A0A067RM58 A0A1Y1KK29 A0A0K8SX23 A0A0K8TEK1 A0A1Y1KSD4 A0A0J7L3V4 A0A1Y1KQ17 A0A1Y1KKC9 A0A1Y1KQ27 A0A0K8SVW5 A0A1Y1KMH7 A0A1B6D953 A0A1B6DM49 A0A1B6EB12 A0A1B6C175 A0A1Y1KK27 E0VZT3 T1HWB4 A0A1Y1KK49 A0A336KXW8 A0A1Q3F495 A0A146KZU4 A0A0K8TF41 A0A0K8T914 A0A0K8SVV8 A0A0K8TET8 A0A1W7R720 Q16XS6 A0A1S4FKD3 A0A2S2P7F6 A0A0K8SVW2 A0A2M3Z2A4 W4VRS5 B0WAD3 A0A336MRU8 A0A1I8N9R0 A0A1I8N9R1 A0A1I8N9Q8 A0A1I8NNX1 A0A1I8N9S0 A0A1B0BMW2 A0A146LW71 A0A1I8NNX9 A0A1I8NNX8 A0A1I8NNX3 A0A1I8NNZ3 A0A336LL25 A0A0A1X0T2 A0A2M4BFH1 W5J694 A0A2M3Z6K3 A0A2M4CG35 A0A2M4AMJ2
Ontologies
GO
PANTHER
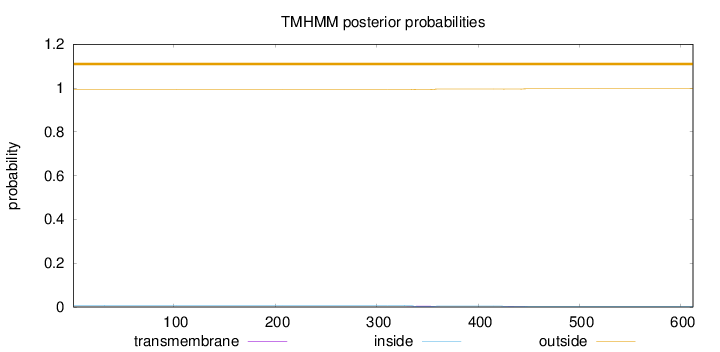
Topology
Length:
612
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16742
Exp number, first 60 AAs:
0.00135
Total prob of N-in:
0.00630
outside
1 - 612
Population Genetic Test Statistics
Pi
1.044689
Theta
2.188746
Tajima's D
-2.162168
CLR
0.2745
CSRT
0.00264986750662467
Interpretation
Possibly Positive selection
